Figure EV3. Mapping of amino acid sequence differences between K1/K5 and K10/K14 onto the molecular surface of the wild‐type K1/K10‐1B structure.
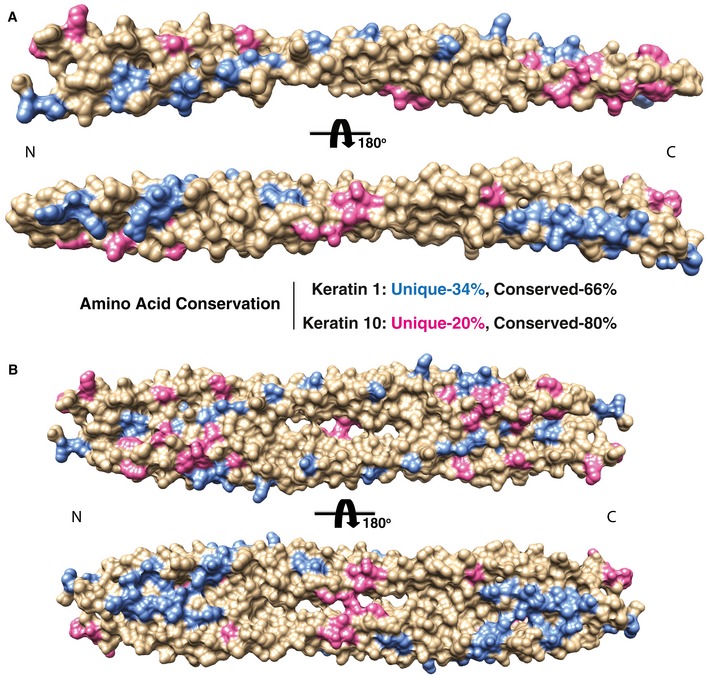
- Residues in K1 (blue) and K10 (pink) that are unique (non‐identical) compared with basal keratins K5 and K14, respectively, are mapped onto the K1/K10‐1B heterodimer structure (represented as a molecular surface). Residues that are conserved/identical are tan. K1 residues that are non‐identical to K5 are colored blue, while K10 residues that are non‐identical to K14 are colored pink. The majority of unique residues for K1 and K10 align along the outer aspect of the helices. The majority of the conserved residues lie along the dimer interface. This distribution of unique residues enables keratins to maximize the diversity of surface properties, such as surface charge and shape, while maintaining a common structural coiled‐coil fold.
- The same mapping and coloring of unique residues as in panel (A) is applied to the K1/K10‐1B tetramer structure.
