Figure 7.
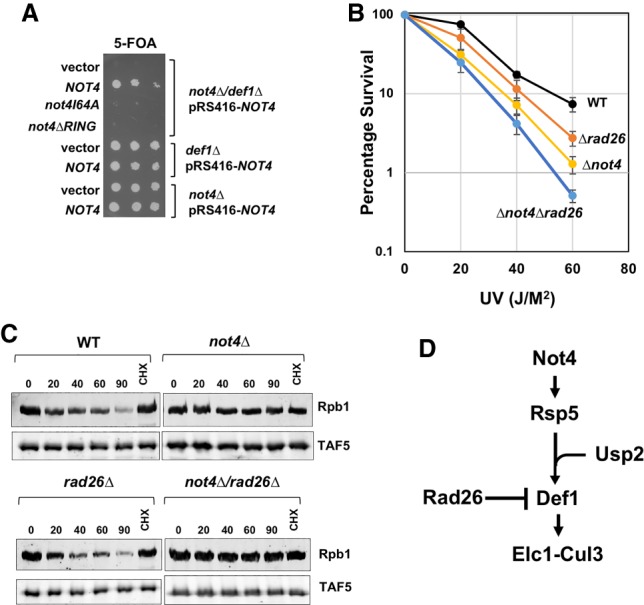
Genetic analysis of NOT4 in the Rpb1 destruction pathway. (A) Plasmid shuffle assay. Single or double DEF1 and/or NOT4 deletion strains containing pRS416-NOT4 (URA3 marked plasmid) were transformed with a LEU2-marked empty vector, one containing wild-type NOT4 or RING domain mutants. Strains were plated onto 5-FOA medium. (B) Epistasis analysis between NOT4 and RAD26. Each point represents the average and standard deviations of three biological replicates. (C) Rpb1 degradation in double mutants. The experiment was performed as described in Figure 1A. The lane labeled “CHX” indicates cells treated with cycloheximide only for 90 min. (D) Proposed role of Not4 in Rpb1 ubiquitylation.
