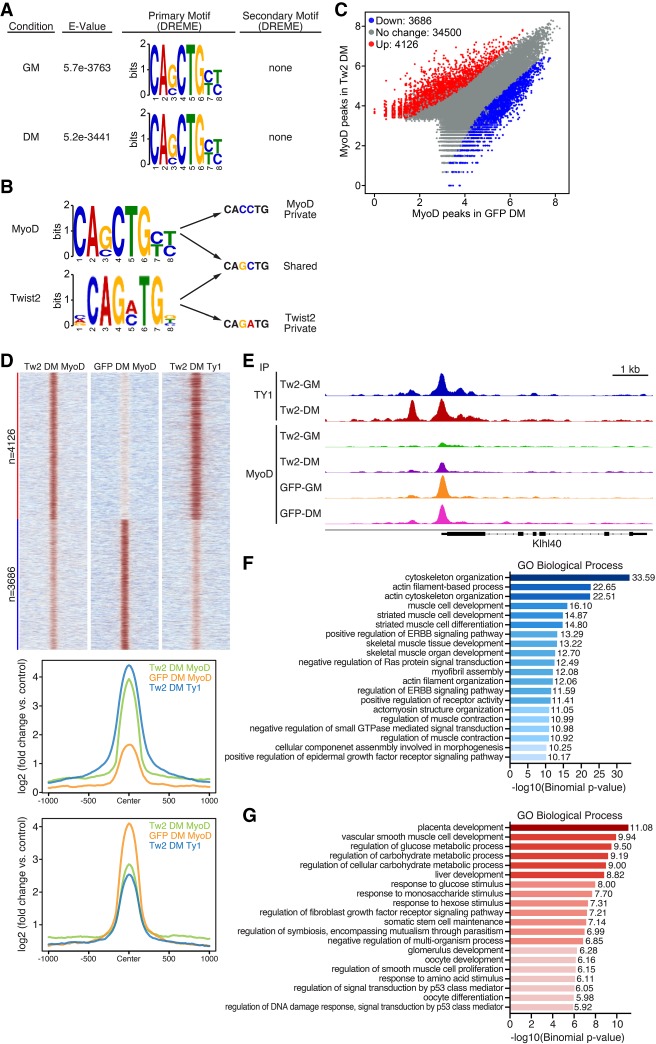
Figure 5.
Twist2 drives global redirection of MyoD binding. (A) De novo motif analysis of the MyoD-binding motif in GM and DM performed by MEME-ChIP. MyoD has no preference for secondary motifs. (B) Comparison of Twist2 and MyoD private and shared E-box variants. (C) Scatter plot of differential MyoD peaks in the presence and absence of Twist2 in DM. Differential peak cutoff was defined as signal twofold or greater change and Padj < 0.05. (D, top) Heat map depicting differential binding of MyoD peaks in the presence or absence of Twist2 in DM. (Bottom) ChIP signal distribution plot of differentially bound MyoD peaks in the presence or absence of Twist2. (E) Genome browser shot depicting Twist2 and MyoD competitive binding at the Klhl40 locus. (F) GREAT (Genomic Region Enrichment of Annotations Tool) analysis of MyoD peaks down-regulated in TW2-DM versus GFP-DM. (G) GREAT analysis of MyoD peaks up-regulated in TW2-DM versus GFP-DM.