Figure 1.
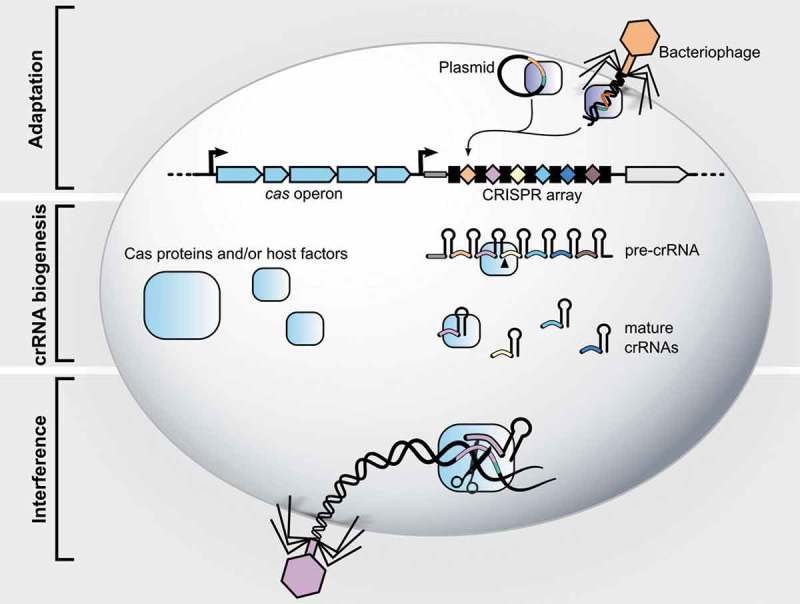
The three stages of CRISPR-Cas adaptive immunity. Stage 1: Adaptation. During this phase, the bacterium incorporates a fragment of the invading phage or plasmid DNA into its genome as a spacer into the CRISPR array (leader: thick grey line; repeats: black boxes; spacers: colored diamonds). This spacer will serve as memory allowing the bacterium to recognize the same threat upon reinfection. After infection, the Cas1-Cas2 complex (dark blue rectangles) recognizes the invading DNA and integrates a portion of it (orange) into the CRISPR array, giving rise to a new spacer (orange diamond). At the same time, a new repeat is generated. Stage 2: crRNA biogenesis. The CRISPR array is transcribed as a long precursor CRISPR RNA (pre-crRNA), which is then processed (as indicated by the black arrow) to generate mature CRISPR RNAs (crRNAs), each containing part of a repeat and of a spacer. Processing is mediated either by a Cas nuclease (Class 1 systems, types V and VI) or a host factor (type II). Stage 3: Interference. Either a complex of Cas proteins or a singular Cas protein guided by the mature crRNAs cleave the nucleic acid target in a sequence-specific manner (as indicated by the scissors). Upon base paring of the crRNA spacer (purple) with the spacer-complementary sequence, known as protospacer (purple), a Cas nuclease catalyzes cleavage of the invading DNA leading to its degradation. The PAM sequence, depicted as a green line, is a short sequence found next to the protospacer but absent in the CRISPR array, which prevents the array to be cleaved by Cas proteins (autoimmunity). Note that blue rectangles can represent a complex of several Cas proteins or a single Cas nuclease depending on the CRISPR class.
