Figure 3.
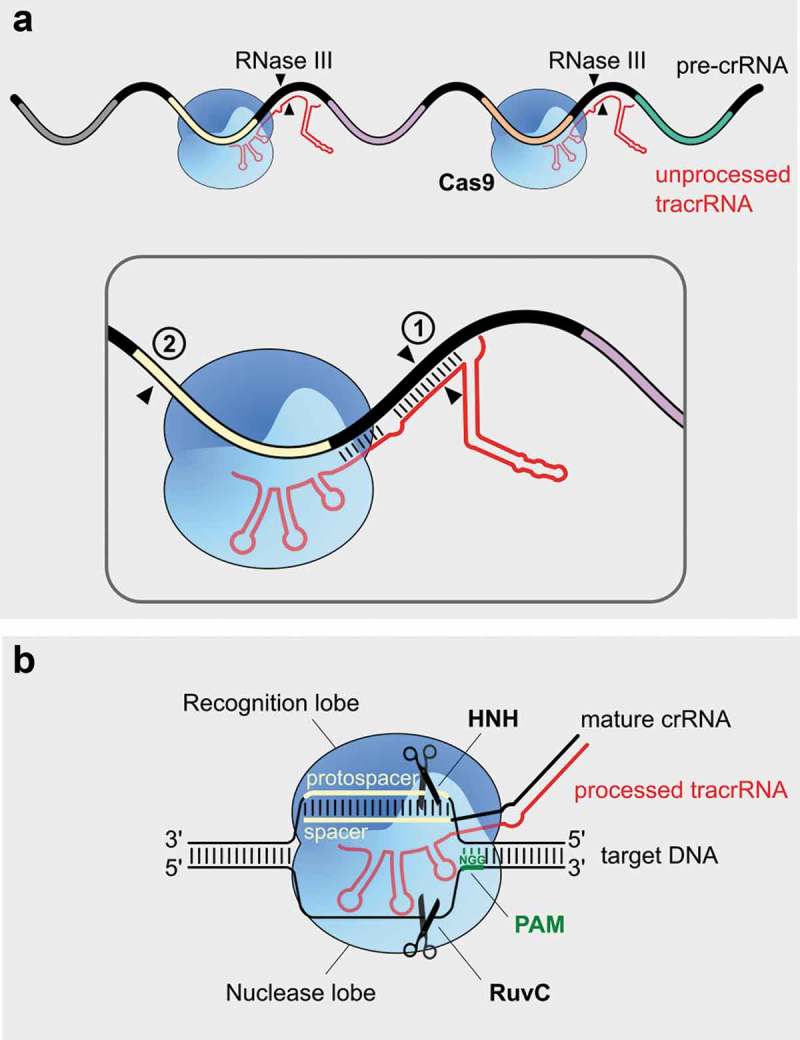
Mechanism of CRISPR-Cas maturation and interference in type II systems. (a). CRISPR RNA maturation by RNase III. The anti-repeat region of tracrRNA (in red) base pairs with the repeats of the pre-crRNA (in black) in a Cas9-dependent manner (blue). ① During the first processing event, the host RNase III recognizes the tracrRNA:crRNA-Cas9 complex and cleaves both tracrRNA and crRNA (depicted with 2 black triangles) within the anti-repeat:repeat duplex. ② A second maturation event (black triangle) takes place in the 5ʹ of crRNA spacer (yellow) by an unknown mechanism, and leads to the production of mature crRNAs. After maturation, the tracrRNA:crRNA duplex remains bound to Cas9. (b). DNA interference by Cas9. The Cas9 endonuclease is then guided by the mature tracrRNA:crRNA duplex to the invading double-stranded DNA (dsDNA). Following Cas9 recognition of the NGG PAM (in green), present immediately downstream of the protospacer on the non-complementary strand, the crRNA spacer and its target DNA (both colored in yellow) base pair and form an R-loop. This triggers cleavage of the DNA by Cas9 nuclease domains (HNH and RuvC, light blue). The HNH domain cleaves the complementary strand, while the RuvC domain cleaves the non-complementary strand, which results in a double strand break on the target DNA. The cleavage takes place three base pairs upstream of the PAM sequence (indicated by the scissors) .
