Figure 2.
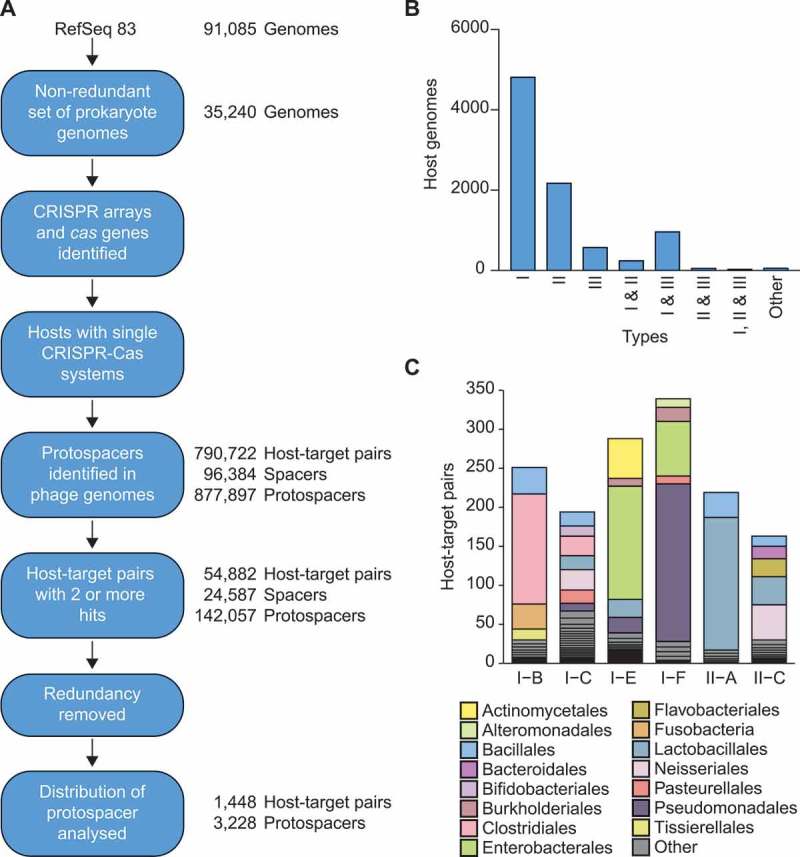
Identification of archaeal virus, phage and prophage protospacers. A) An overview of the steps used to generate the non-redundant host-target dataset of spacer-protospacer matches. For details see Methods. B) The number of genomes possessing each designated CRISPR-Cas type or multiple co-occurring types, based on the cas genes identified. Due to their low occurrences, all other types and combinations are included in ‘other’ (i.e. types IV, V and VI along with any other subtype co-occurrences). C) Taxonomic overview of the final dataset. Species-level diversity is shown in Fig. S1.
