Figure 5.
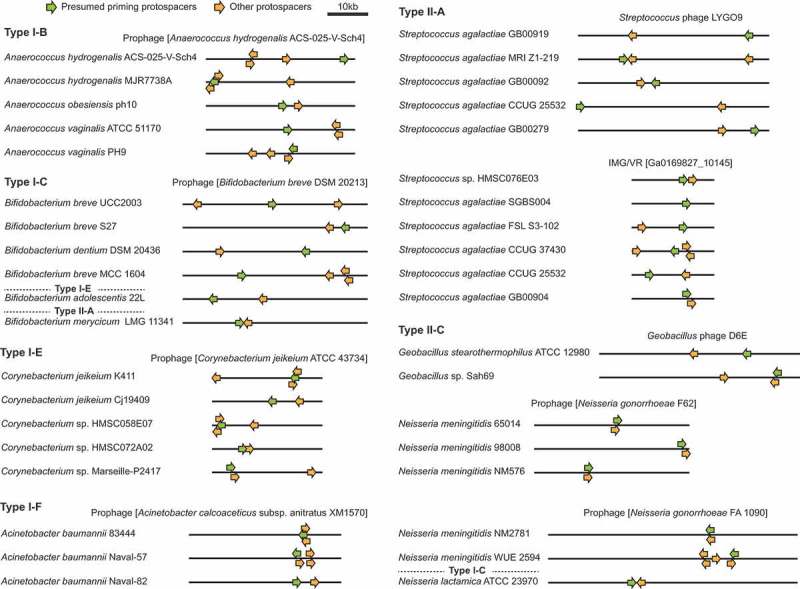
Protospacer clustering on shared target genomes reveals primed CRISPR adaptation. Protospacer clustering for sets of closely-related host species targeting the same phages or prophages. Note that protospacers toward either end of the displayed linear phage genome are considered clustered because most phage genomes undergo circularised states during replication. The IMG/VR accessions represent viral contigs assembled from metagenomic data [59]. In some cases the displayed host species are representative of several related hosts that possess the same spacer sets matching the specified phage. Examples of single hosts with spacers matching multiple targets are provided in Fig. S5.
