Figure 2.
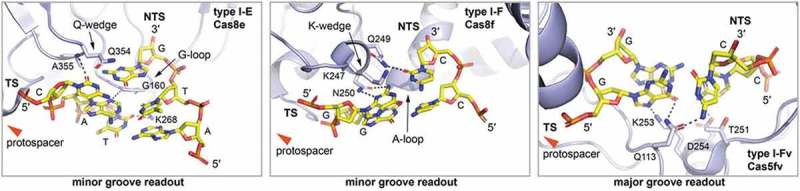
PAM recognition by type I Cascades. Left: Close up on the PAM interacting region of the E. coli type I-E Cascade subunit Cas8e (light blue) (PDB: 5H9E [75]). Cas8e promiscuously recognizes the ATG PAM (yellow) via a set of polar interactions (black dashed lines) from the DNA minor groove. The glycine rich loop (G-loop) recognizes the second base pair of the PAM. The Q-wedge might assist in target strand (TS) protospacer displacement from the non-target strand (NTS) protospacer complementary sequence. The red arrow indicates the direction of the protospacer. Middle: Close up on the PAM interacting region of the P. aeruginosa type I-F Cascade subunit Cas8f (light blue) (PDB: 6B44 [38]). Cas8e specifically recognizes the GG PAM (yellow) via a set of polar interactions (black dashed lines) from the DNA minor groove. The alanine rich loop (A-loop) recognizes the second base pair of the PAM. The K-wedge might assist in target strand (TS) protospacer displacement from the non-target strand (NTS) protospacer complementary sequence. The red arrow indicates the direction of the protospacer. Right: Close up on the PAM interacting region of the S. putrefaciens type I-Fv Cascade subunit Cas5fv (light blue) (PDB: 5O6U [82]). Cas5fv specifically recognizes the GG PAM (yellow) via a set of polar interactions (black dashed lines) from the DNA major groove. Base pairing of the first PAM GC base pair is distorted by Q113. The red arrow indicates the direction of the protospacer.
