Figure 5.
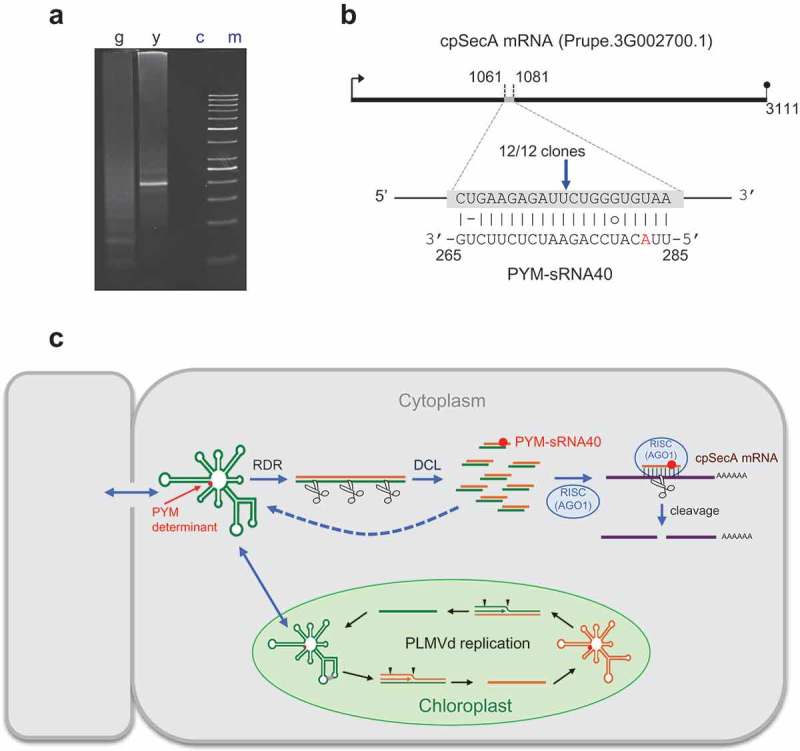
Site-specific cleavage of peach cpSecA mRNA guided by PYM-sRNA40. (a) RLM-RACE of two RNA preparations from a GF-305 peach seedling inoculated with PLMVd variant y4, one from fully green (g) leaves and the other from yellow (y) sectors of PYM-expressing leaves; (c) refers to a control without added template and (m) to the size markers. PAGE of the nested RT-PCR products reveals, only in the yellow sectors, a band of the expected size (195 bp), essentially co-migrating with the 200-bp DNA marker. (b) Duplex formed between the cpSecA mRNA and PYM-sRNA40; notice that the latter derives from the PLMVd (−) strand, with the nucleotide strictly associated with PYM (position 283 in PLMVd variant y4) highlighted in red. Arrow points to the predicted cleavage site (within the sixth exon) validated by RLM-RACE, with the fraction showing the number of independent clones producing identical results. (c) Model for the genesis of the PYM-sRNA40 (in the cytoplasm) that also includes PLMVd replication (within chloroplasts, with arrowheads marking hammerhead self-cleavage sites). RDR and DCL refer to RNA-directed RNA polymerase(s) and dicer-like ribonuclease(s), respectively. Green and yellow lines denote PLMVd (+) and (−) strands, respectively. PLMVd-sRNAs likely target the genomic viroid RNAs for their RISC(AGO)-mediated cleavage (with some fragments being transformed by RDR into additional DCL substrates); PYM-sRNA40 guides RISC(AGO1) for cleaving the cpSecA mRNA. Red dot indicates the PYM-associated nucleotide.
