Figure 8.
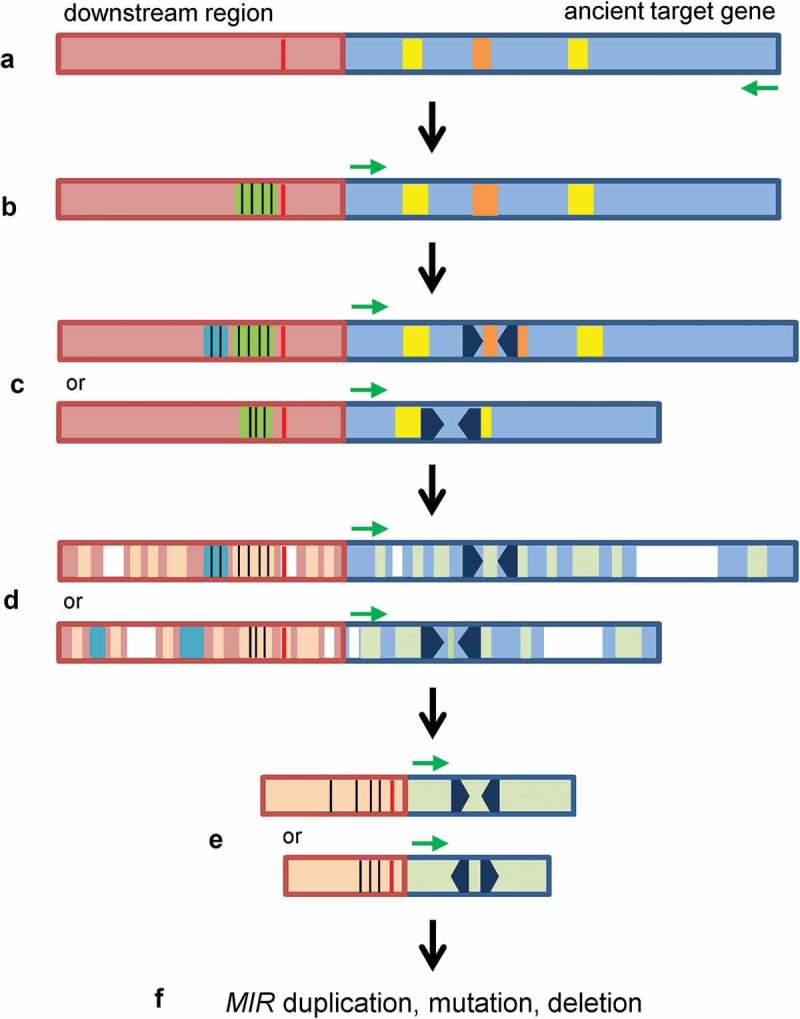
The short inverted repeat generation model for de novo origination and evolution of plant MIRs. (A) An ancient target gene (light blue) with palindromic-like (brown) and/or imperfect IR (yellow) sequences. Transcription direction is indicated by a green arrow. AT-rich TATA-box-like sequence is shown in red. (B) A MITE sequence (green) with significant cis-elements (black vertical lines), such as CAAT-box and CuREs, was inserted in the flanking region of the ancient target gene or a copy of the duplicated target gene. The insertion of MITE and subsequent evolution resulted in the gain of promoter activity. (C) Short inverted repeats (black arrowheads) were generated in the palindromic-like or imperfect IR sequences. Cis-elements could be dynamically changed (blue) in the promoters. (D) Deletion (white), insertion (blue) and mutation (light brown and light green) occurred during evolution. (E) Evolved MIRs. (F) The evolved MIRs could expand through duplication, diversify by mutation or lose by deletion during plant evolution.
