Figure 1.
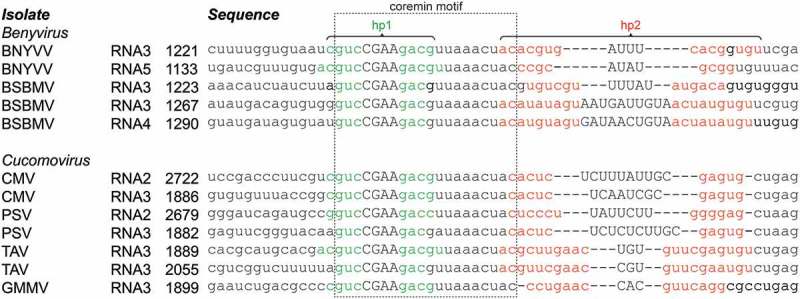
Alignment of coremin motifs in beny- and cucumoviruses. Multiple beny- and cucomovirus species harbor a coremin motif (boxed), which carries nucleotides that form a putative 3–5 bp-sized hairpin structure (green; uppercase letters depict the predicted loop sequence). An additional putative hairpin, more variable than, and directly downstream of the coremin motif, has been predicted by MFOLD for each sequence (base pairs formed by red nucleotides). Through structural alignment, covariations are revealed in this region. The position of the 5ʹ most nt is indicated for each sequence. RNA3 of BSBMV and TAV isolates harbor two proximate coremin motifs. Note that this list is not exhaustive but shows the variation within these two genera. BNYVV: Beet necrotic yellow vein virus (RNA3: KX665538, RNA5: KP316977), BSBMV: Beet soil-borne mosaic virus (RNA3: KX352171, RNA4: KX352034), CMV: Cucumber mosaic virus (RNA2: KX013371, RNA3: KX013372), PSV: Peanut stunt virus (RNA2: MF170158, RNA3: AY775057), TAV: Tomato aspermy virus (L79971), GMMV: Gayfeather mild mottle virus (FM881901).
