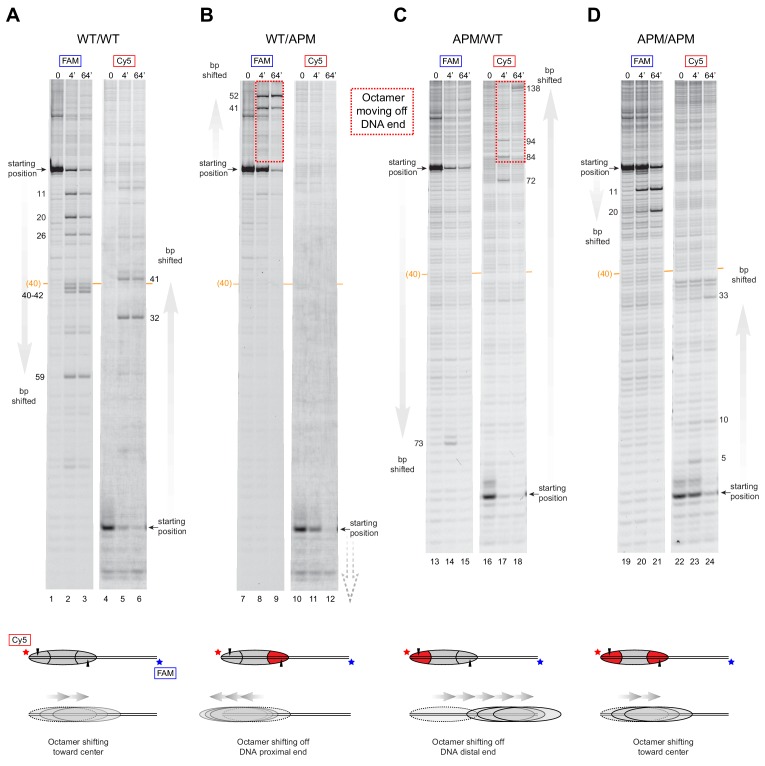
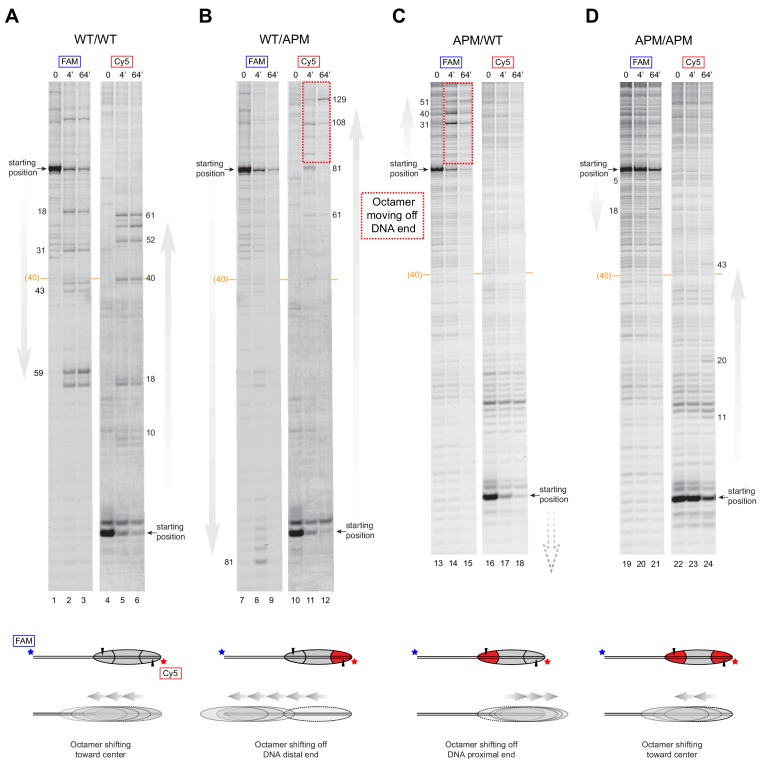
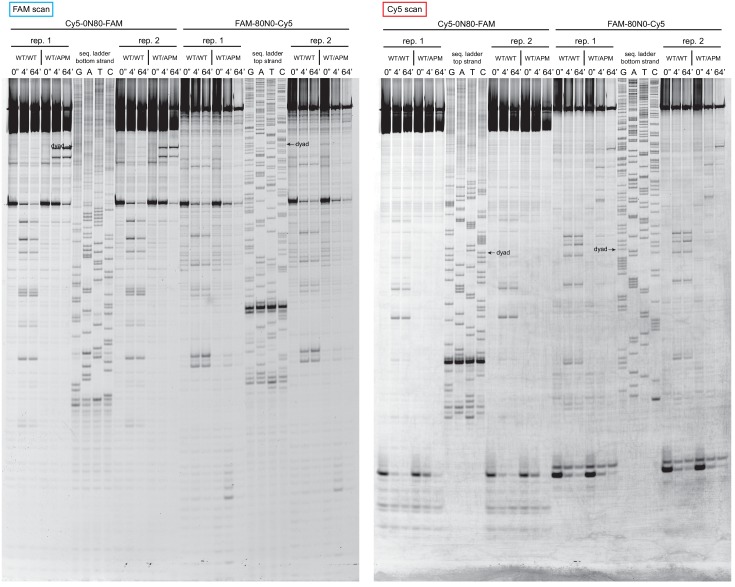
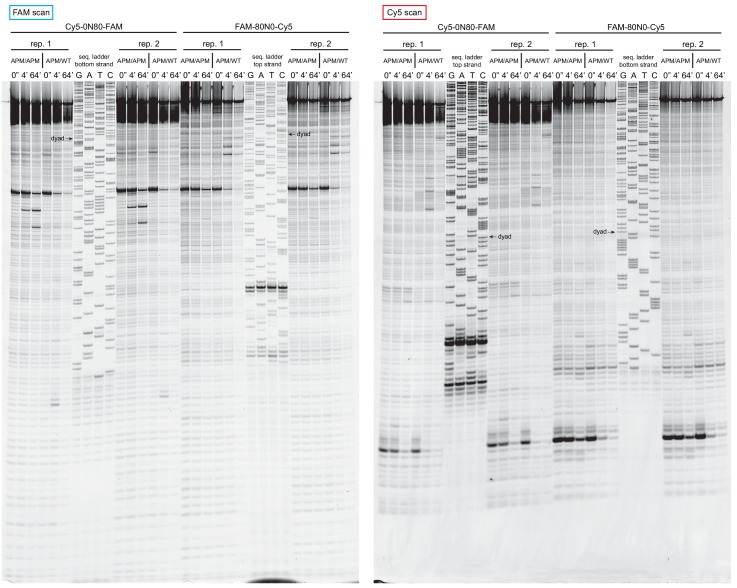
Figure 4. SNF2h slides histone cores with asymmetric acidic patch mutations off DNA ends.
Histone mapping of SNF2h sliding reactions, performed with 0N80 nucleosomes having the four combinations of wild type and APM H2A/H2B dimers: (A) WT/WT, (B) WT/APM, (C) APM/WT, and (D) APM/APM. Shown are scans of urea denaturing gels, which report on the locations of the histone octamer based on sites of H2B(S53C) cross-linking. Two scans are shown for each nucleosome, based on Cy5 and FAM labels on the top and bottom DNA strands, respectively. Numbering beside the gels indicates the distances (bp) the histone octamer shifted relative to the starting position, with the midpoint of the 80 bp flanking DNA highlighted with a central orange bar. Each nucleosome was generated by addition of 200 nM wild type (gray) or APM (red) H2A/H2B dimers to 100 nM wild type or APM hexasome. Cartoon schematics below each gel indicate the composition and orientation of APM and wild type H2A/H2B dimers, the sites of H2B(S53C) cross-linking (black triangles), and interpretations of sliding reactions. Sites where the histone octamer moves off DNA ends are highlighted by dotted red boxes. Sliding reactions contained 1 µM SNF2h and 2 mM ATP, and reactions were quenched at 0, 4, and 64 min. These results are representative of two independent experiments. Analogous experiments using 80N0 nucleosomes are shown in Figure 4—figure supplement 1. Extended gels are shown in Figure 4—figure supplement 2 and Figure 4—figure supplement 3.