Figure 2.
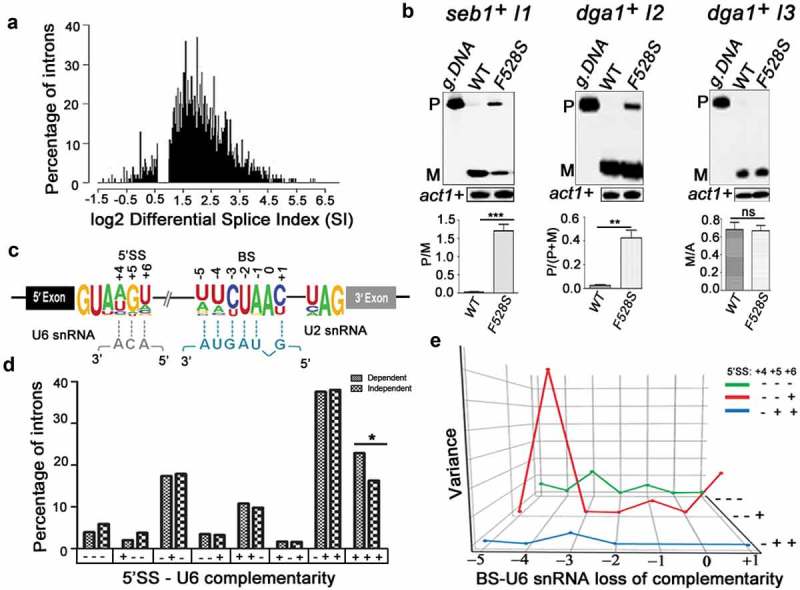
SpPrp16 has nearly ubiquitous splicing functions. (a) Frequency histogram representing the number of introns plotted against their respective log2 differential splice index (SI). Introns with log2 differential SI ≥ 1 (i.e., ≥2 fold difference between the wild-type and mutant) are considered strongly dependent on SpPrp16 for splicing and those with log2 differential SI < 0.6 (i.e. <1.5 fold difference between the wild-type and mutant) are considered unaffected in the spprp16F528S mutant. The introns with marginally affected differential SI values (log2 differential splice index between 0.6 and 1) were excluded from this representation. (b) Semi-quantitative RT-PCR validation of the transcriptome-based splicing phenotype at 30°C for SpPrp16 dependent seb1+ intron1 (left panel), dga1+ intron2 (middle panel) and independent intron dga1+ intron3 (right panel). (c) Schematic of a S. pombe transcript with global sequence consensus for the intronic 5’SS, BS and 3’SS elements. Base-pairing of the 5’SS and the BS with U6 snRNA and U2 snRNA, respectively, are indicated by dotted lines. (d) Graph representing the number of introns from SpPrp16 dependent and independent classes categorized based on the complementarity of 5’SS +4, +5 and +6 nucleotides with the U6 snRNA sequence – ACA. Base pairing at each position is denoted by a ‘+’sign and no base pairing by ‘–’ sign. The number of introns with complete complementarity indicated as ‘+++’ is significantly different between the dependent and independent intron categories at p = 0.03. (e) Principal Component Analysis (PCA) showing variance between the number of dependent and independent introns (Y axis) with loss of BS-U2 snRNA base-pairing for each indicated BS residue (X axis). The lines (Z axis) represent data for intron subsets with varying 5’SS-U6 snRNA strength. Green (– – –), red (– – +) and blue (– + +) lines indicates three intron subsets categorized based on 5’SS-U6 complementarity at the 5’SS +4, +5 and +6 positions. The invariant branch residue A was positioned as ‘0’ and other BS nucleotides are numbered with respect to this residue.
