Figure 6.
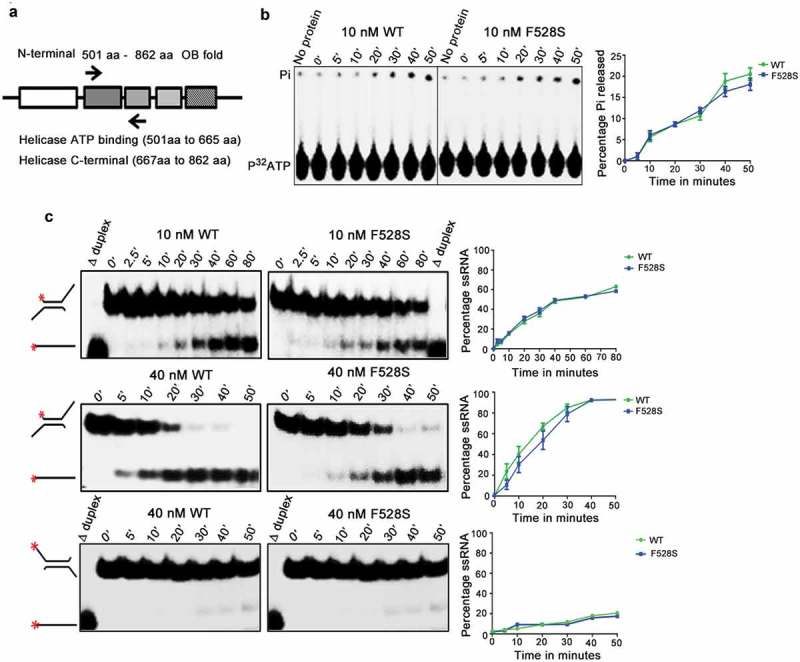
Biochemical characterization of Spprp16 helicase domain mutants. (a) Schematic of SpPrp16 domain architecture. Arrow marks indicate primer positions used to amplify the helicase domain (501–862 amino acids) of SpPrp16 wild type and mutant proteins. (b) ATP hydrolysis by wild-type and Spprp16F528S helicase protein upon incubating 10 nM of each protein with 1mM ATP and tracer amounts of γP32ATP at 30°C for the different time points indicated. (c) dsRNA unwinding activity of the wild-type and Spprp16F528S helicase proteins on a 3′ overhang containing RNA duplex labelled at the 5′ end (schematic to the left). The reactions were arrested at the indicated time points (in minutes). The upper and middle panels represent the activity of 10 nM and 40 nM proteins, respectively. Δ duplex indicates heat denatured RNA duplex, a marker for ssRNA. Bottom panel shows helicase activity of the wild-type and Spprp16F528S helicase protein on a 5ʹ overhang containing RNA duplex; 40 nM of wild-type and mutant protein was used in this experiment.
