Figure 7.
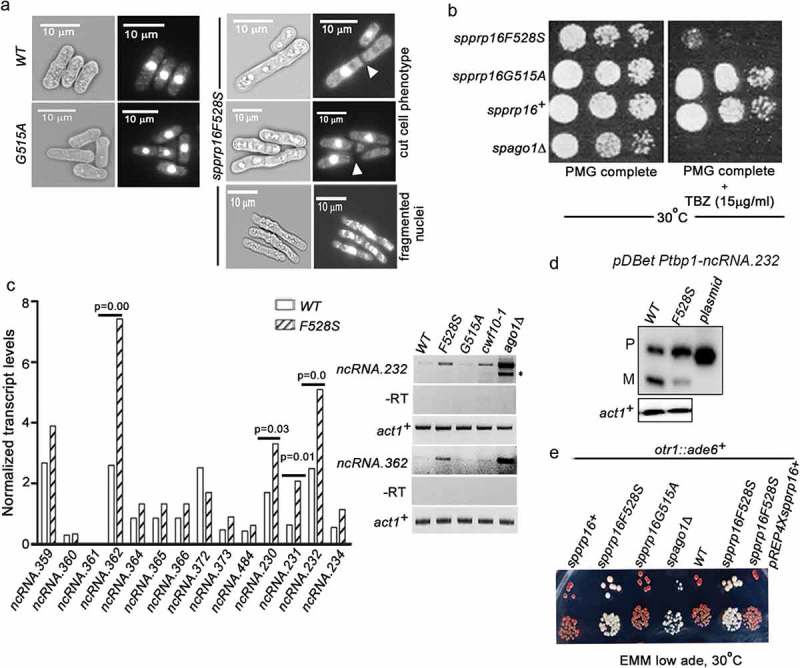
SpPrp16 is required for successful mitosis and for efficient transcriptional gene silencing at centromeres and telomeres. (a) Bright field and DAPI stained images of spprp16+, spprp16F528S and spprp16G515A cells. The ‘cut’ cells (white arrowheads at the site of cell separation) and lagging chromosome (fragmented nuclei) in spprp16F528S are shown. Scale bar is 10 µm in all panels. (b) Thiabendazole (TBZ) sensitivity of spprp16 mutants as compared to wild-type and spago1Δ strains at 30°C. Cultures were grown to equivalent OD595 before spotting 10-fold serial dilutions on PMG complete media containing TBZ (at 15 µg/ml concentration). The growth proficiency on PMG complete media (left) served as the control. (c) Normalized transcript levels for centromeric ncRNAs obtained from RNA-seq of spprp16+ (WT) and spprp16F528S mutant cells (left graph). P value significance for upregulated ncRNAs in the spprp16F528S mutant is shown. The semi-quantitative RT-PCR for two centromeric transcripts ncRNA.232 and ncRNA.362 are shown in the right upper and lower panels respectively. For ncRNA.232, ‘*’ to the right side of the gel indicates the unspliced precursor. Control PCR reactions without reverse transcription are shown in the -RT panel. (d) Semi-quantitative RT-PCR to assess the splicing of ncRNA.232 mini-transcript in the WT (leu1:spprp16+) and F528S (leu1:spprp16F528S) mutant strain. (e) Transcriptional silencing of ade6+ centromeric reporter in the wild-type and spprp16F528S mutant cells assessed in media with limiting adenine. cwf10-1 and spago1Δ served as positive controls for defective centromeric heterochromatin.
