ABSTRACT
CRISPR-Cas defends microbial cells against invading nucleic acids including viral genomes. Recent studies have shown that type III-A CRISPR-Cas systems target both RNA and DNA in a transcription-dependent manner. We previously found a type III-A system on a conjugative plasmid in Lactococcus lactis which provided resistance against virulent phages of the Siphoviridae family. Its naturally occurring spacers are oriented to generate crRNAs complementary to target phage mRNA, suggesting transcription-dependent targeting. Here, we show that only constructs whose spacers produce crRNAs complementary to the phage mRNA confer phage resistance in L. lactis. In vivo nucleic acid cleavage assays showed that cleavage of phage dsDNA genome was not detected within phage-infected L. lactis cells. On the other hand, Northern blots indicated that the lactococcal CRISPR-Cas cleaves phage mRNA in vivo. These results cannot exclude that single-stranded phage DNA is not being targeted, but phage DNA replication has been shown to be impaired.
KEYWORDS: Phages, CRISPR, Lactococcus, resistance
Introduction
Lactococcus lactis is an industrial bacterium primarily used for the production of fermented dairy products. Virulent bacterial viruses (phages), commonly found in the manufacturing environment, can cause economic loss due to end-product quality issues and failed fermentation processes. Research into phage-L. lactis interactions has revealed an extensive repertoire of distinct phage defense mechanisms. The genetic basis for many of these naturally occurring systems has been identified, furthering our understanding of the evolution of phage-host interactions and enabling their exploitation in strain construction for improved phage resistance [1,2].
CRISPR-Cas (clustered regularly interspaced short palindromic repeats – CRISPR associated proteins) is a potent phage defense mechanism and has been found in several bacteria and archaea. This adaptive immune system consists of short repeat sequences interspaced by short variable/spacer sequences derived from invading nucleic acids. CRISPR-Cas systems generally function in three steps: 1) adaptation, where new spacers are incorporated into the CRISPR array; 2) maturation or processing, during which crRNAs are generated to guide the Cas protein machinery towards their respective nucleic acid targets; 3) interference, in which the crRNA targeted invader nucleic acid is cleaved and/or degraded [3]. Depending on specific effector and accessory modules, CRISPR-Cas systems are classified into two classes, with multi-subunit effector complexes in Class 1 and single-protein effector modules in Class 2 [4]. These systems are further divided into six types (Types I – VI) and into several subtypes (designated by a letter).
L. lactis was thought to be devoid of CRISPR-Cas systems, instead largely relying on other defense systems such as phage adsorption blocking, superinfection immunity, restriction-modification, and abortive infection mechanisms [5]. While very effective, these defenses do not afford the adaptability of CRISPR-Cas. A novel type III-A CRISPR-Cas was recently discovered in an industrial L. lactis strain [6]. The system was found in only five strains out of > 400 strains examined [6]. The lactococcal CRISPR-Cas system is encoded on a 62.8-kb conjugative plasmid and provides resistance against phages targeted by native spacers [6]. The utility of phage resistance and natural transferability prompted further investigations into functioning of the Type III-A system and its practical application for lactococcal phage resistance.
Extensive studies of homologous type III CRISPR-Cas systems have provided insights into its mechanism of action [7]. For type III systems, nucleic acid targeting is transcription-dependent, meaning that the crRNA (encoded by the CRISPR array) must be complementary to the target mRNA for degradation to occur [8,9]. In addition, the interference machinery of type III-A systems generally includes cas6 which is involved in crRNA processing, endoribonuclease csm6, and five Csm genes (csm1/cas10, csm2-csm5) which make up the effector complex [10]. It is the Csm3 nuclease that cleaves target RNA [11,12]. Interaction of the Csm complex with the target RNA also leads to DNA degradation by Csm1/Cas10. Additionally, recent studies have shown that the binding of target RNA by the Csm complex results in the synthesis of cyclic oligoadenylates by Cas10 which activate non-specific RNA degradation by Csm6 [13,14].
Very recently, the lactococcal type III-A CRISPR-Cas system was expressed in Escherichia coli and shown to display transcription-dependent plasmid DNA interference [10]. While studying spacer requirements for phage resistance in the native lactococcal CRISPR-Cas, we also observed that spacers conferring phage interference were oriented such that their expected crRNAs would be complementary to the target phage mRNA; consistent with transcription-dependent targeting.
In this study, we investigated the fate of phage dsDNA and mRNA after the infection of lactococcal cells carrying the native Type III-A CRISPR-Cas system in which the CRISPR array was engineered with various spacers transcribed in both orientations.
Results and discussion
The native lactococcal CRISPR-Cas system previously described in strain L. lactis DGCC7167 contains 15 spacers, varying from of 33 to 38-bp in size, of which spacers S2, S3, and S4 were shown to provide phage resistance [6]. As oriented in the CRISPR array, all three spacers match the non-coding strand of their respective protospacer target in the phage genomes (Table 1). Based on the direction of transcription of the CRISPR locus, each spacer would lead to crRNAs complementary to the protospacer mRNA (Fig. 1 and Table 1).
Table 1.
Select pKLM spacers and random spacer nucleotide sequence, relevant properties, and efficiency of plaquing on phages P008, bIL170, and P335.
| Spacer | Phage | ntID | Length(nt) | Sequence (5' - 3') | Protospacer strand | Efficiency of plaquing | ||
|---|---|---|---|---|---|---|---|---|
| P008 | bIL107 | P335 | ||||||
| None | - | - | - | - | 1 | 1 | 1 | |
| pKLM S2 | P008 | 33 | 35 | TTTTTAAAATGTTGCAAATGTTTAGCTACTTCAT | NC | 4.4 x 10−6 | - | - |
| pKLM S3 | P008 | 28 | 35 | TTCTGTTAATTTAACTCCCATTTGTTAGTTCTCCT | NC | <9.3 x 10−8 | - | - |
| pKLM S4 | bIL170 | 30 | 35 | ATACGTTCTTTGAACCAAGCTTCAACTCCCTCGGA | NC | |||
| P008 | 31 | NC | 4.5 x 10−7 | 2.7 x 10−6 | - | |||
| RS3n | P335 | 36 | 36 | ATGCAGTGCGTTGCCAATGATATACTGTTACTGAACCAATTACT | NC | ≤1 x 10-8 | - | 8.1 x 10−5 |
| RS3c | P008 | 36 | GGTTCAGCAGTAATTGGTTCAGTAACAGTATATCATTGGCAACG | C | 1.1 | - | 1.1 | |
| RS4n | bIL170 | 39 | 39 | AGTTACACCATTTTCAGCAAGTTTTGCGTTCCAAGCGTTTTTAATTT | NC | ≤1 x 10−8 | ≤1 x 10−7 | - |
| RS4c | P008 | 39 | AAAATCAGAAATTAAAAACGCTTGGAACGCAAAACTTGCTGAAAATG | C | 1.1 | 1.3 | - | |
| RS9n | bIL170 | 35 | 35 | CAGGCACTACGATTGTTCCGTCATTATTACCCTTTGTAGCAGT | NC | ≤1 x 10−8 | ≤1 x 10−7 | - |
| RS9c | P008 | 35 | ATGTAACTACTGCTACAAAGGGTAATAATGACGGAACAATCGT | C | 1.1 | 1.2 | - | |
| RS21n | bIL170 | 35 | 35 | TCTTCGTCAAACTTTTCTAGCTCTTTGATTAATTCTTTTACTT | NC | 2.1 x 10−4 | 3.4 x 10−4 | - |
| RS21c | P008 | 35 | WARAATGAAAGTAAAAGAATTAATCAAAGAGCTAGAAAAGTTT | C | 0.9 | 1 | - | |
| RS36n | P008 | 35 | 35 | TTATTTCCATCTATATGGTCAACGGTTAAATCACTTTTACCGT | NC | 1.7 x 10−6 | - | - |
| RS36c | AGCTTTTTACGGTAAAAGTGATTTAACCGTTGACCATATAGAT | C | 1 | - | - | |||
| RS53n | bIL170 | 31 | 35 | TTCAACAKCATTAAGTAAGAACTCGTTAAAGTCAACCATTGAC | NC | 6.4 x 10−7 | 4.1 x 10−6 | - |
| RS53c | P008 | 35 | GAAACATTGTCAATGGTTGACTTTAACGAGTTCTTACTTAATG | C | 1.1 | 0.9 | - | |
C, Coding; NC, Non-coding; (-) Not applicable; Nucleotides in Bold and Underlined represent 5’ flanking regions
Figure 1.
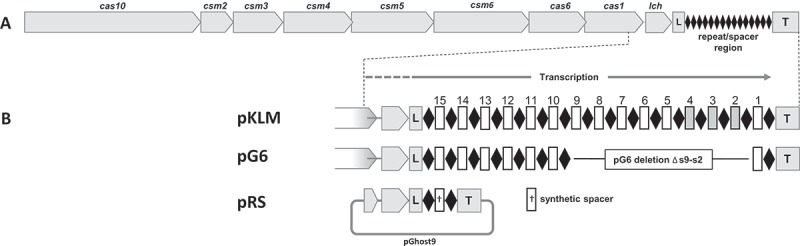
Lactococcal CRISPR-Cas synthetic spacer interference.
(A) Type III-A CRISPR-Cas operon from pKLM. CRISPR-associated (cas) genes are represented as grey arrows oriented towards the direction of transcription. The leader (L) and trailer (T) sequences are shown as boxes with the repeat-spacer (CRISPR) array depicted as black diamonds.(B) Expanded view of the repeat-spacer array and flanking regions. Repeats are shown as black diamonds and spacers as rectangles. Plasmids pKLM and pG6 have a complete set of cas genes. The pKLM array has the full complement of spacers whereas pG6 array lacks spacers S2 through S9. Spacers S2, S3, and S4 have nucleotide identity and provide resistance to phages P008 and bIL170. To construct random spacer delivery vector pRS, the region demarcated by dashed lines was cloned into the broad host range vector pGhost9 and contains the 3ʹ end of cas1, lch, leader, and trailer sequence flanking a single repeat-spacer-repeat unit.
To confirm that this orientation bias is a condition for phage resistance, we introduced phage-targeting spacers into the CRISPR array in both orientations (protospacer coding or non-coding strand) and tested their ability to confer in vivo phage resistance. Six randomly selected spacers (denoted ‘RS’ in Fig. 1 and Table 1) were designed and synthesized from the lactococcal strictly virulent phage P008, a member of the Siphoviridae family with a double-stranded DNA (dsDNA) genome of 28,538 bp [15]. Random spacers were selected from regions of P008 genome which are conserved among various phages in order that single spacers could be used against multiple phages. All spacers were chosen so that the 5ʹ flanking region of the protospacer would not have homology with the 8 nucleotide-tag (5ʹ-ACGAGAAC-3ʹ) on the crRNA as homology in this region has been shown to provide CRISPR-Cas immunity [9]. Because the native spacers in the lactococcal array vary in size, our synthetic spacers varied from 35 – 39 bp in length. The synthetic spacers were a perfect sequence match to various sections of the phage P008 genome and target early, middle or late expressed genes [16]. The synthetic spacers were assembled as a single repeat-spacer-repeat array flanked by sequences immediately proximal to the lactococcal CRISPR, including the leader. The single spacer arrays with their flanking sequences were cloned into the lactococcal vector pGhost9 [17] (EmR erythromycin resistance; spacer vector constructs denoted as ‘pRS’) and individually transformed into the phage-sensitive strain L. lactis 1403S containing plasmid pG6 [6]. Plasmid pG6 is an erythromycin sensitive variant of the lactococcal conjugative plasmid pKLM encoding the type III-A CRISPR-Cas system with a truncated CRISPR array. The pG6 array lacks pKLM spacers S9 – S2 which direct interference against virulent phage P008 and also against related lytic phages p2 and bIL170 (Fig. 1). Randomly selected transformants, each containing one of the various pRS plasmids, were confirmed by sequencing and tested for resistance to phages.
Phage resistance was only observed in transformants harboring spacers matching the respective phage protospacer non-coding strand (Table 1). The lactococcal type III-A system provided a resistance phenotype ranging from a 4-log reduction in plaquing efficiency to complete inhibition (> 8 log reduction; no plaque formation) (Table 1), depending on the spacers. In addition to P008, sequence identity of the designed spacers was also found against phages bIL170 (Sk1virus genus, formely type 936) and/or P335 (eponymous type P335) that infect L. lactis 1403S (Table 1). The same resistance phenotype was observed indicating the response is not phage specific. The array in pRS constructs which did not confer interference (pRS3c, pRS4c, pRS9c, pRS21c, pRS36c, pRS53c) was sequenced and confirmed to be in the expected orientation and maintained an intact RS insert sequence. These results show that a crRNA complementary to the protospacer mRNA is a condition for phage interference which is consistent with the transcription-dependent targeting phenotype previously described for the heterologously expressed lactococcal CRISPR-Cas as well as other type III-A CRISPR-Cas systems [8–12].
To observe the in vivo fate of phage mRNA, RNA cleavage assays were developed using the virulent lactococcal phage P008. The pRS constructs which target orf53 of phage P008 were selected for the RNA cleavage assays as the RS53 protospacer resides in a genomic region that generates a single transcript [16]. L. lactis strains IL1403, 1403S-RS53n, and 1403S-RS53c were individually infected with phage P008, and total RNA was extracted from a non-infected cell sample as well as from phage-infected cells at 0, 5, 10, 15, 20 and 30 minutes post-infection. Then, Northern blots were performed with probes designed to detect either side of the orf53 protospacer on an expected transcript of 1.1-kb. mRNA cleavage would result in fragments of 1-kb and approx. 0.05-kb. No phage mRNA cleavage was observed for control phage-infected L. lactis strains IL1403 or 1403S-RS53c, consistent with their phage sensitivity phenotypes. For the phage-infected and phage-resistant strain L. lactis 1403S-RS53n, a < 0.06-kb cleavage product was clearly visible and increased over time; likewise, the probe for the 1-kb fragment (Fig. 2) also detected cleavage and degradation. RNA cleavage assays were repeated using RS36n and RS36c, which target orf36 of phage P008. RS36 results corroborate the results from RS53 (Fig. 3). We concluded that the lactococcal CRISPR-Cas cleaved the targeted phage mRNA when a spacer was correctly oriented within the array.
Figure 2.
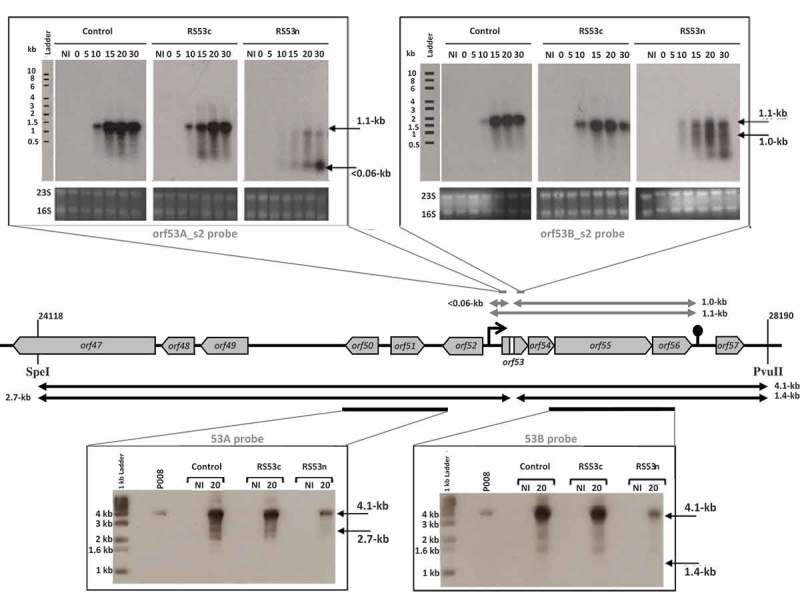
The lactococcal CRISPR-Cas targets phage RNA; orf53.
The middle section depicts the relevant segment of the virulent lactococcal P008 genome analyzed for the cleavage assays. In the upper panel, arrows and Northern blot show the corresponding 1.1-kb phage mRNA transcript and subsequent fragments (< 0.06-kb and 1.0-kb) that would be generated if the mRNA is targeted. In the bottom panel, arrows and Southern blot show the phage genomic DNA 4.1-kb fragment generated by SpeI-PvuII digestion and expected fragments of 2.7-kb and 1.4-kb if DNA is targeted. Equivalent amounts of DNA digest was loaded for each lane (see Materials & Methods and Supplementary Figure 1). The diminished signal intensity for the R53n, where there is phage inhibition, is a function of the amount of P008 phage DNA relative to total DNA in the extraction and digestion. The gray bars indicate the position of the probes used to detect the respective fragments. The bacterial strains used for the phage infection were L. lactis 1403S (phage-sensitive control), L. lactis 1403S-RS53c, and L. lactis 1403S-RS53n all infected or not by the strictly lytic phage P008. NI = not infected. The time in minutes post phage infection is indicated on each autoradiogram.
Figure 3.
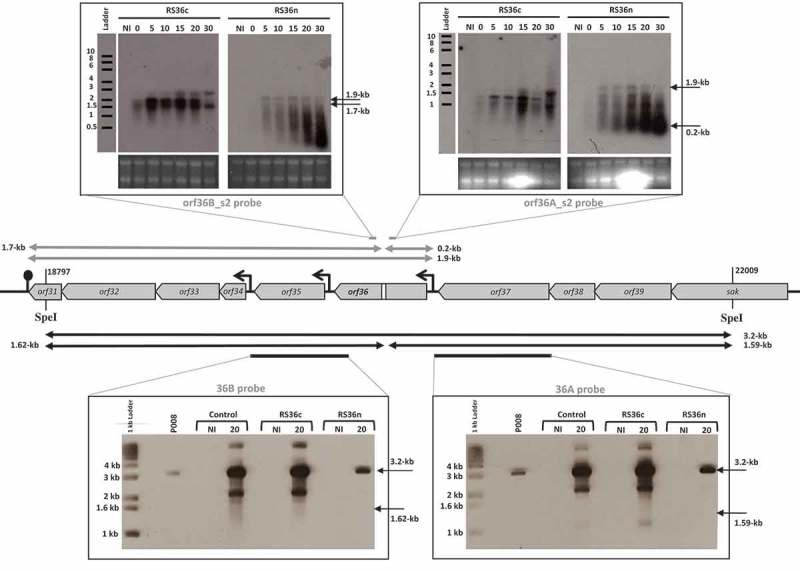
The lactococcal CRISPR-Cas targets phage RNA; orf36.
The middle section depicts the relevant segment of the virulent lactococcal P008 genome for the cleavage assays. In the upper panel, arrows and Northern blot show the corresponding 1.9-kb phage mRNA transcript and subsequent fragments (0.2-kb and 1.7-kb) that would be generated if mRNA is targeted. In the bottom panel, arrows and Southern blot show the phage genomic DNA 3.2-kb fragment generated by SpeI digestion and expected fragments of 1.62-kb and 1.59-kb if DNA is targeted. Equivalent amounts of DNA digest was loaded for each lane (see Materials & Methods and Supplementary Figure 1). The diminished signal intensity for the R36n, where there is phage inhibition, is a function of the amount of P008 phage DNA relative to total DNA in the extraction and digestion. The gray bars indicate the position of the probes used to detect the respective fragments. The strains used for the infection were L. lactis 1403S (phage-sensitive control), L. lactis 1403S-RS36c, and L. lactis 1403S-RS36n all infected or not by the strictly lytic phage P008. NI = not infected. The time in minutes post phage infection is indicated on each autoradiogram.
To detect cleavage of phage dsDNA genome, L. lactis IL1403, 1403S-RS53n, and 1403S-RS53c were infected with phage P008, and total DNA was extracted from non-infected cell sample as well as from phage-infected cells 20 minutes post-infection. Southern blots were performed on total DNA extracts digested with SpeI and PvuII which generated a 4.1-kb fragment that includes the orf53 target (Fig. 2). Probes were designed to detect either side of the orf53 protospacer in the phage P008 genome if the CRISPR-Cas mediated double-stranded DNA cleavage as observed for type II-A systems [18,19]. Such cleavage of the phage genome within or in the vicinity of the targeted protospacer would result in dsDNA fragments of 2.7-kb and 1.4-kb. Only the intact 4.1-kb frament was observed indicating that no cleavage of phage dsDNA was detected in our assay. DNA cleavage assay was repeated using RS36n and RS36c. RS36 results corroborate the results from RS53 (Fig. 3).
The lack of observed dsDNA cleavage is in agreement with recent mechanistic studies showing that the type III-A effector complexes of S. thermophilus, S. epidermidis, and T. thermophilus, when bound to an mRNA transcript, cleave single-stranded DNA (ssDNA) and not dsDNA [9,12,20]. The reduction in total phage P008 DNA seen in the Southern blots in cells containing active spacers RS36n or RS53n is consistent with the phage resistance phenotype and possibly ssDNA targeting (Figs. 2 & 3). CRISPR-Cas interference reduces phage propagation leading to a decrease in total phage DNA as compared to the control where phages are actively replicating (Fig. 4). The proposed model for ssDNA targeting involves the binding of the Csm complex to the target mRNA which then allows for the cleavage of ssDNA within the transcription bubble [14,20]. This ssDNAse functionality resides within the Cas10 protein in type III-A systems [9,14,20,21].
Figure 4.
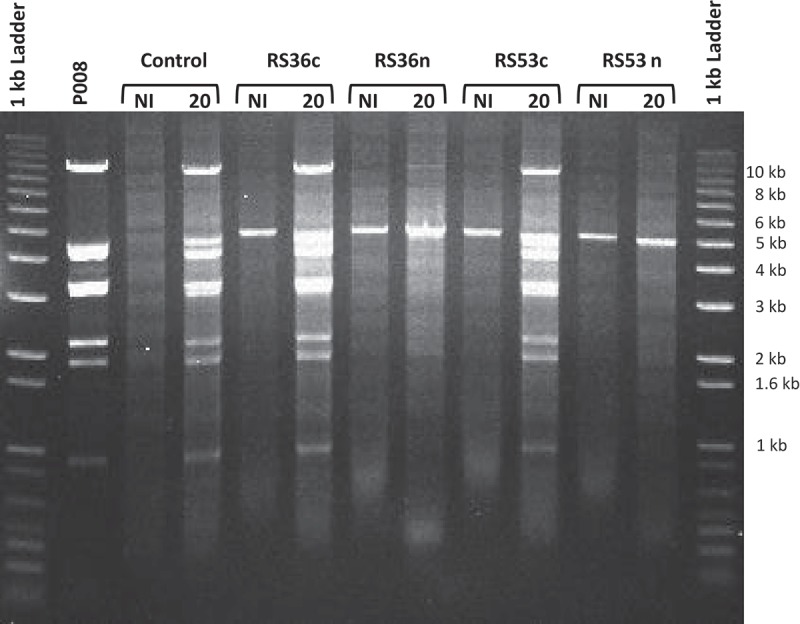
The lactococcal CRISPR-Cas interfere with phage DNA replication; orf36 and orf53.
Migration of the total DNA extractions, obtained from phage P008-infected and non-infected L. lactis strains as well as phage P008 only, digested with SpeI and PvuII restriction enzymes. The respective strains used for the infection were L. lactis 1403S (control), L. lactis 1403S-RS36c, L. lactis 1403S-RS36n, L. lactis 1403S-RS53c, and L. lactis 1403S-RS53n. The prominent banding pattern in phage infected lanes represents replication of phage DNA over host genomic DNA as a consequence of phage infection. The 5-kb band is the pRS vector which is at higher copy than host genomic DNA and contains a single SpeI site. The time 20 minutes post phage infection was used and is indicated when applicable. NI = not infected.
Protection against virulent phages is a fundamental necessity for bacterial survival. For L. lactis, a bacterium extensively used in industrial applications where phages are prevalent, the ability to defend itself is critical. The results presented here demonstrate that the lactococcal type III-A CRISPR-Cas system provide phage resistance when the CRISPR array is engineered with spacers oriented to generate crRNAs complementary that target phage mRNA. The availability of a naturally occurring, self-transmissible, interference functional, lactococcal CRISPR-Cas offers a means to develop robust phage resistant industrial strains [6].
In comparison to type II systems, it has been suggested that type III-A system afforded a more robust phage immunity due in part to the lack of a PAM (protospacer adjacent motif) sequence requirement, tolerance for nucleotide mismatches within the target sequence (protospacer), and effectiveness of inhibition from a single spacer, especially when targeting essential genes [22]. Genetic similarities between the L. lactis type III-A CRISPR and other type III-A systems would predict similar function. We demonstrated that the in vivo functionality of the lactococcal type III-A system is in general agreement with the mechanism of action of similar systems found in S. epidermidis [8,9], S. thermophilus [20], and Thermus thermophilus [12]. The flexibility in spacer targeting and strength of single spacer derived immunity, especially when targeting essential genes, offer significant advantage towards constructing industrial strains with enhanced and programmable phage resistance.
Materials and methods
Bacterial strains, bacteriophages, and plasmids
The biological materials used in the study are listed in Table 2. All L. lactis strains were grown at 30°C in M17 broth (Becton, Dickenson and Co.) supplemented with 0.5% glucose (Glu). Escherichia coli was propagated aerobically in LB broth (Becton, Dickenson and Co.) at 37°C. When required, antibiotics were added to the media as follows: Erythromycin (Em) 5 µg/ml for lactococci or 150 µg/ml for E. coli; spectinomycin (Sp) 300 µg/ml, streptomycin (Sm) 1000 µg/ml. Preparation of bacteriophage lysates and standard plaque assays were performed as described [23]. Efficiency of plaquing results are the average of at least 3 independent assays.
Table 2.
Strains, plasmids, and bacteriophages.
| Strains | Description | Reference |
|---|---|---|
| E. coli TG1 RepA+ | supE hsd∆5 thi ∆(kac-proAB) F’ (traD36 proAB+ lacIq lacZ∆M15) with chromosomal copy of pWV01 repA | [30] |
| L. lactis IL1403 | Laboratory strain | [31] |
| L. lactis 1403S | Spontaneous SmR derivative of IL1403 | [6] |
| L. lactis 1403S-RS3c | 1403S + pG6 containing pRS3c | This work |
| L. lactis 1403S-RS3n | 1403S + pG6 containing pRS3n | This work |
| L. lactis 1403S-RS4c | 1403S + pG6 containing pRS4c | This work |
| L. lactis 1403S-RS4n | 1403S + pG6 containing pRS4n | This work |
| L. lactis 1403S-RS9c | 1403S + pG6 containing pRS9c | This work |
| L. lactis 1403S-RS9n | 1403S + pG6 containing pRS9n | This work |
| L. lactis 1403S-RS21c | 1403S + pG6 containing pRS21c | This work |
| L. lactis 1403S-RS21n | 1403S + pG6 containing pRS21n | This work |
| L. lactis 1403S-RS36c | 1403S + pG6 containing pRS36c | This work |
| L. lactis 1403S-RS36n | 1403S + pG6 containing pRS36n | This work |
| L. lactis 1403S-RS53c | 1403S + pG6 containing pRS53c | This work |
| L. lactis 1403S-RS53n | 1403S + pG6 containing pRS53n | This work |
| Plasmids | ||
| pG6 | pKLM CRISPR-Cas ∆s2-s9 | [6] |
| pGhost9 | EmR, TS ori Temperature sensitive vector | [17] |
| pKLM | CRISPR-Cas | [6] |
| pRS3c | pGhost9 + RS3c | This work |
| pRS3n | pGhost9 + RS3n | This work |
| pRS4c | pGhost9 + RS4c | This work |
| pRS4n | pGhost9 + RS4n | This work |
| pRS9c | pGhost9 + RS9c | This work |
| pRS9n | pGhost9 + RS9n | This work |
| pRS21c | pGhost9 + RS21c | This work |
| pRS21n | pGhost9 + RS21n | This work |
| pRS36c | pGhost9 + RS36c | This work |
| pRS36n | pGhost9 + RS36n | This work |
| pRS53c | pGhost9 + RS53c | This work |
| pRS53n | pGhost9 + RS53n | This work |
| Phages | ||
| bIL170 | Host L. lactis IL1403/1403S | [32] |
| P008 | Host L. lactis IL1403/1403S | [15] |
| P335 | Host L. lactis IL1403/1403S | [33] |
PCR reactions
PCR was performed with Phusion HF Mastermix (New England Biolabs) according to manufacturer’s instructions. Primers were synthesized by Integrated DNA Technologies, and primer sequences are listed in Table 3. PCR products were purified for Gibson assembly or sequencing using a NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel). Sequencing of PCR amplicons was performed by Eurofins MWG Operon, and results were analyzed using Clustal Omega (Sievers).
Table 3.
List of primers used in the study.
| Primer Name |
Sequence 5ʹ – 3ʹ |
|---|---|
| GB-VF1194 | CATGTTCAATACGGACACGTA |
| GB-VR | AAGCTTTTCCGCGCTATTGT |
| InsFspacer | TTCTGGAATGGGATTCAGGATTGTTGATGTGTTGGTATTTGAG |
| InsRspacer | ATCCGTGTCGTTCTGTCCACTACGTGTCCGTATTGAACATG |
| orf53A_s1 | GCAGAAGAACAGCTACTATTTAAGCAAGAAAC |
| orf53A_s2 | GTTTCTTGCTTAAATAGTAGCTGTTCTTCTG |
| orf53B_s1 | GCTGTTGAATGTGGTTTGATTAATCTTGATACAGC |
| orf53B_s2 | GCTGTATCAAGATTAATCAAACCACATTCAACAG |
| orf36A_s1 | GCCGGGTTAACAAAAACAAAATCTGGTTATTTAAGAG |
| orf36A_s2 | GCTCTTAAATAACCAGATTTTGTTTTTGTTAACCCGG |
| orf36B_s1 | GTAGCTAAATACGTCGGAGTTACCCAGCCAG |
| orf36B_s2 | GCTGGCTGGGTAACTCCGACGTATTTAGCTA |
| 936.101F | GCGAAGCCAACACCTACAAT |
| 936.56R | GCTAAATACGTCGGAGTTAC |
| 936.60F | AGGAGTTTCTGGTTCGTCCC |
| 936.47R | TAGGCGACACCATAACGGTA |
| 936.94F | CTAAGCCCTCTAATTCTCCT |
| 936.36R | CACGCTTGACCGCGATTTAA |
| 936.34F | GAGCAAAGCCTTAGCGATTG |
| 936.92R | GCTAGATAGTCATGGCGATA |
| SpacerF | Forward RS sequence + AAATACAACCGCTCCTCGATAAAAGGGGACGAGAACCATATGATTCAGGTATTGC Depending on spacer size, the 3ʹ end was shortened to obtain a primer of 90 nt |
| SpacerR | Reverse complement RS sequence + GTTCTCGTCCCCTTTTATCGAGGAGCGGTTGTATTTAGAGAACTTTAAAAACGTG Depending on spacer size, the 3ʹ end was shortened to obtain a primer of 90 nt |
| VF-pG9 | GTGGACAGAACGACACGGAT |
| VR-pG9 | TCCTGAATCCCATTCCAGAA |
Clonings
The first set of random spacer cloning inserts was created by overlapping PCR. PCRs were performed with L. lactis 1403S cells containing pG6 using primer set InsFspacer and SpacerR and primer set InsRspacer and SpacerF. Purified amplicons were mixed 1:1 and used as a template in a PCR reaction with primers InsFspacer and InsRspacer. A linear amplicon of vector pGhost9 was created by PCR with primers VF-pG9 and VR-pG9. Cloning inserts were assembled with linear pGhost9 using Gibson Assembly Mastermix (New England Biolabs) according to manufacturer’s instructions. The additional random spacer and cleavage model cloning inserts were ordered as gblocks from Integrated DNA Technologies. PCR was performed on recombinant plasmid pGhost9 + RS9 with primers GB-VF1194 and GB-VR which created a linear vector with the region containing the RS9 deleted. gblocks were assembled with linear vector using Gibson Assembly Mastermix according to manufacturer’s instructions. All Gibson assemblies were electroporated into E. coli TG1 RepA+ cells according to method of Dower [24]. Recombinant plasmids were isolated from TG1 RepA+ cells using QIAprep Spin MiniPrep Kit (Qiagen). Purified plasmids were electroporated into lactococci [25].
RNA extraction and northern blots
The phage infections were done as described elsewhere [26] with the virulent phage P008 and its host L. lactis 1403S containing either plasmid pG6 (control) or the plasmid pG6 containing the CRISPR locus corresponding to the protospacer orf53 or orf36 in (C) coding or in (NC) non-coding strand. Samples were taken at time NI (non-infected), 0, 5, 10, 15, 20 and 30 minutes post-infection and treated with lysozyme (60 mg/ml) for 10 minutes at 37°C to increase bacterial lysis. RNA was extracted with the RNeasy kit as indicated by the supplier (Qiagen), treated with DNase I (Roche) to remove residual DNA, and protected with RNA inhibitor (Roche). The RNA sample concentration was determined with a Nanodrop 2000 (Thermo-Fisher). Five μg of RNA were migrated on 1% denaturing formaldehyde-agarose gels and transferred on nylon membranes [27]. The Northern blot experiments were performed as described previously [28] with 32P oligo-radiolabeled probes (orf53A_s1, orf53A_s2, orf53B_s1, orf53B_s2, orf36A_s1, orf36A_s2, orf36B_s1, orf36B_s2). The A probes correspond to those upstream of the protospacer in the phage genome and the B probes correspond to those downstream. The s1 probes were used to detect potential anti-sense transcripts and the probes s2 the sense transcripts. Loading controls represent the rRNA (23S and 16S) detected directly on the ethidium bromide colored agarose gels.
DNA extraction and southern blots
L. lactis strains were incubated at 30°C in 90 mL of GM17-CaCl2 until they reached an OD600 of 0.5. After removing a 10 mL sample of uninfected cells (NI), each bacterial culture was infected with CsCl-purified phage P008 at a multiplicity of infection of 5 and incubated at 30°C. Samples were taken after 20 min, centrifuged for 5 min at 16,000 g, and pellets were flash frozen and stored at −80°C until DNA extraction. Total DNA extractions were performed simultaneously as described elsewhere [29]. The DNA sample concentration was determined with a Nanodrop 2000 (Thermo-Fisher). Four micrograms of DNA from a phage-infected L. lactis strains and 1 μg of P008 phage were double digested with SpeI and PvuII restriction enzymes (Roche) before migration on a 1.5% agarose gel. Replicated versions of gel migration (one per probe) were done using 1/10 of these DNA amounts (fig.S1). The DNA was then transferred onto positively charged nylon membrane by capillary blotting [27]. One nanogram of digested phage DNA was used as positive control for probe hybridizations. DNA probe similarly targeting the orf53 (53A and 53B) or orf36 (36A and 36B) were prepared with PCR DIG labeling mix (Roche) using the primers 936.94F and 936.36R (53A), 936.34F and 936.92R (53B), 936.60F and 936.47R (36A) and finally 936.101F and 936.56R (36B). Pre-hybridization, hybridization, washes and detection (CDP-star) were performed as recommended by Roche.
Funding Statement
This work was supported in part by the Canada Research Chairs; Natural Sciences and Engineering Research Council of Canada.
Acknowledgments
Authors would like to thank Philippe Horvath and Patrick Boyaval for discussion. S.M. acknowledges funding from the Natural Sciences and Engineering Research Council of Canada. S.M. holds a Tier 1 Canada Research Chair in Bacteriophages.
Disclosure statement
A.M.M, D.A.R., G.M.R. and S.M. are co-inventors on patent(s) or patent application(s) related to CRISPR–Cas systems and their various uses. The remaining authors declare no competing interests.
Supplemental data
Supplemental data for this article can be accessed here.
References
- 1.Samson JE, Moineau S.. Bacteriophages in food fermentations: new frontiers in a continuous arms race. Annu Rev Food Sci Technol. 2013;4:347–368. [DOI] [PubMed] [Google Scholar]
- 2.Mahony J, Bottacini F, van Sinderen D, et al. Progress in lactic acid bacterial phage research. Microb Cell Fact. 2014;13(Suppl 1):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makarova KS, Wolf YI, Alkhnbashi OS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13(11):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horvath P, Coûté-Monvoisin AC, Romero DA, et al. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int J Food Microbiol. 2009;131(1):62–70. [DOI] [PubMed] [Google Scholar]
- 6.Millen AM, Horvath P, Boyaval P, et al. Mobile CRISPR/Cas-mediated bacteriophage resistance in Lactococcus lactis. PLoS ONE. 2012;7(12):e51663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pyenson NC, Marraffini LA. Type III CRISPR-Cas systems: when DNA cleavage just isn’t enough. Curr Opin Microbiol. 2017;37:150–154. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg GW, Jiang W, Bikard D. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014;514:633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samai P, Pyenson N, Jiang W, et al. Co-transcriptional DNA and RNA cleavage during type III CRISPR-Cas immunity. Cell. 2015;161:1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ichikawa HT, Cooper JC, Lo L, et al. Programmable type III-A CRISPR-Cas DNA targeting modules. PLoS ONE. 2017;12(4):e0176221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamulaitis G, Kazlauskiene M, Manakova E, et al. Programmable RNA shredding by the type III-A CRISPR-cas system of Streptococcus thermophilus. Mol Cell. 2014;56:506–517. [DOI] [PubMed] [Google Scholar]
- 12.Staals RHJ, Zhu Y, Taylor DW, et al. RNA targeting by the type III-A CRISPR-cas csm complex of Thermus thermophilus. Mol Cell. 2014;56:518–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kazlauskiene M, Kostiuk G, Venclovas C, et al. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science. 2017;357:605–609. [DOI] [PubMed] [Google Scholar]
- 14.Niewoehner O, Garcia-Doval C, Rostøl JT, et al. Type III CRISPR–cas systems produce cyclic oligoadenylate second messengers. Nature. 2017;548:543–548. [DOI] [PubMed] [Google Scholar]
- 15.Mahony J, Deveau H, Mc Grath S, et al. Sequence and comparative genomic analysis of lactococcal bacteriophages jj50, 712, and P008: evolutionary insights into the 936 phage species. FEMS Microbiol Lett. 2006;261:253–261. [DOI] [PubMed] [Google Scholar]
- 16.Samson JE, Bélanger M, Moineau S. Effect of the abortive infection mechanism and type III toxin/antitoxin system AbiQ on the lytic cycle of Lactococcus lactis phages. J Bacteriol. 2013;195:3947–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maguin E, Prévost H, Ehrlich SD, et al. Efficient insertional mutagenesis in lactococci and other Gram-positive bacteria. J Bacteriol. 1996;178:931–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garneau JE, Dupuis M-E, Villion M, et al. The CRISPR/Cas bacterial immune system cleaves phage and plasmid DNA. Nature. 2010;468:67–71. [DOI] [PubMed] [Google Scholar]
- 19.Magadán AH, Dupuis M-E, Villion M, et al. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas System. PLoS ONE. 2012;7(7):e40913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazlauskiene M, Tamulaitis G, Kostiuk G, et al. Spatiotemporal control of Type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol Cell. 2016;62:295–306. [DOI] [PubMed] [Google Scholar]
- 21.Hatoum-Aslan A, Maniv I, Samai P, et al. Genetic characterization of antiplasmid immunity through a type III-A CRISPR-Cas system. J Bacteriol. 2014;196:310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pyenson NC, Gayvert K, Varble A, et al. Broad targeting specificity during bacterial type III CRISPR-Cas immunity constrains viral escape. Cell Host & Microbe. 2017;22:343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moineau S, Fortier J, Ackermann HW, et al. Characterization of lactococcal bacteriophages from Québec cheese plants. Can J Microbiol. 1992;38:875–882. [Google Scholar]
- 24.Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E.coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holo H, Nes IF. High-frequency transformation, by electroporation of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samson JE, Spinelli S, Cambillau C, et al. Structure and activity of AbiQ, a lactococcal endoribonuclease belonging to the type-III toxin-antitoxin system. Mol Microbiol. 2013;87:756–768. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Russell RW. Molecular cloning: A laboratory manual. 3rd ed. Cold Spring Harbor, (NY): Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 28.Fortier LC, Bransi A, Moineau S. Genome sequence and global gene expression of Q54, a new phage species linking the 936 and c2 phage species of Lactococcus lactis. J Bacteriol. 2006;188:6101–6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fortier LC, Moineau S. Morphological and genetic diversity of temperate phages in Clostridium difficile. Appl Environ Microbiol. 2007;73:7358–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolotin A, Wincker P, Mauger S, et al. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp.lactis IL1403. Genome Res. 2001;11:731–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duwat P, Cochu A, Ehrlich SD, et al. Characterization of Lactococcus lactis UV-sensitive mutants obtained by ISS1 transposition. J Bacteriol. 1997;179:4473–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crutz-Le Coq AM, Cesselin B, Commissaire J, et al. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology. 2002;148:985–1001. [DOI] [PubMed] [Google Scholar]
- 33.Labrie SJ, Josephsen J, Neve H, et al. Morphology, genome sequence, and structural proteome of type phage P335 from Lactococcus lactis. Appl Environ Microbiol. 2008;74:4636–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


