Figure 2.
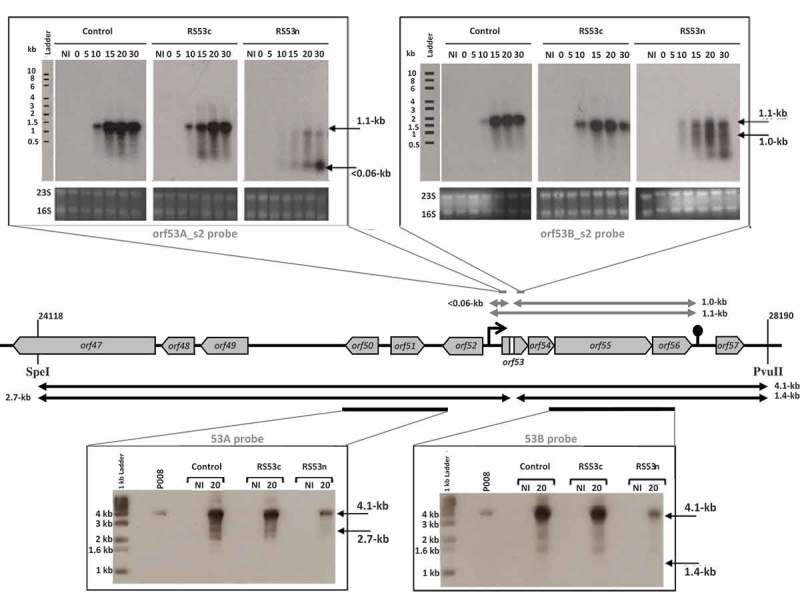
The lactococcal CRISPR-Cas targets phage RNA; orf53.
The middle section depicts the relevant segment of the virulent lactococcal P008 genome analyzed for the cleavage assays. In the upper panel, arrows and Northern blot show the corresponding 1.1-kb phage mRNA transcript and subsequent fragments (< 0.06-kb and 1.0-kb) that would be generated if the mRNA is targeted. In the bottom panel, arrows and Southern blot show the phage genomic DNA 4.1-kb fragment generated by SpeI-PvuII digestion and expected fragments of 2.7-kb and 1.4-kb if DNA is targeted. Equivalent amounts of DNA digest was loaded for each lane (see Materials & Methods and Supplementary Figure 1). The diminished signal intensity for the R53n, where there is phage inhibition, is a function of the amount of P008 phage DNA relative to total DNA in the extraction and digestion. The gray bars indicate the position of the probes used to detect the respective fragments. The bacterial strains used for the phage infection were L. lactis 1403S (phage-sensitive control), L. lactis 1403S-RS53c, and L. lactis 1403S-RS53n all infected or not by the strictly lytic phage P008. NI = not infected. The time in minutes post phage infection is indicated on each autoradiogram.
