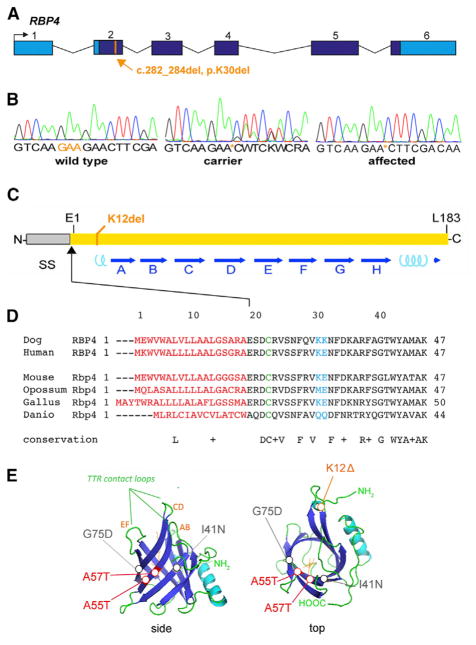
Figure 3. Pathogenic RBP4 Deletion.
(A) p.K30del mutation. Genomic map shows the solitary coding variant identified in the critical region by whole genome sequencing, a 3-bp deletion in exon 2 that removes lysine codon 30 from the RBP precursor. This residue corresponds to K12 in the mature polypeptide, after signal peptide cleavage. Coding (dark blue) and UTR sequences (light blue) are indicated.
(B) Sanger chromatograms showing the DNA sequence of PCR products spanning the RBP4 deletion in wild-type (WT), carrier, and affected dogs. The deletion removes one of two tandem lysine codons (AAG).
(C) Linear diagram of RBP showing the signal peptide (SS); 8 antiparallel β sheets (A–H, blue arrows), which form the ligand barrel; 2 short α-helical segments (cyan coils); 3 cysteine disulfide bonds, which stabilize the tertiary structure; and the mutated K12 residue.
(D) Alignment of vertebrate RBP sequences showing evolutionary conservation of K12 among eutherians. The signal peptide (red), tandem lysines (K12–K13, blue), and disulfide-linked cysteine (C4, green) are indicated. The N-terminal segment preceding the β-barrel (10 of 21 residues) is highly charged. (E) Tertiary structure of canine RBP (ribbon views), modeled from human apo (1RBP) and holo (1BRQ) RBP X-ray data (Cowan et al., 1990; Zanotti et al., 1993a, 1993b), showing K12 near the N terminus, within an α-helical region. By shortening this segment, K12del may limit apposition of C4 and C160 side groups in the ER, preventing formation of one disulfide bond in vivo and, consequently, destabilizing the protein.
See also Table S2.