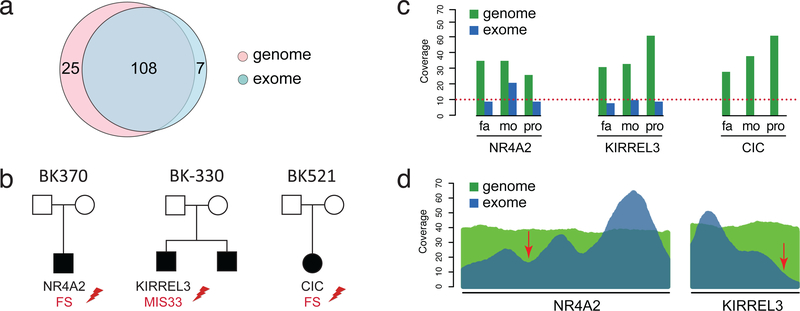
Figure 1. GS versus ES detection.
(a) Comparison of the number of de novo coding SNVs and indels detected in exome-targeted regions by GS and ES. (b) Families with de novo variants in NDD risk genes missed by ES. (c) Comparison of ES and GS coverage of variants in NR4A2, KIRREL3, and CIC in mother, father, and proband. (d) Mean coverage across all samples sequenced by both GS and ES across NR4A2 and KIRREL3. Arrows point to location of the two variants discovered by GS but missed in ES.