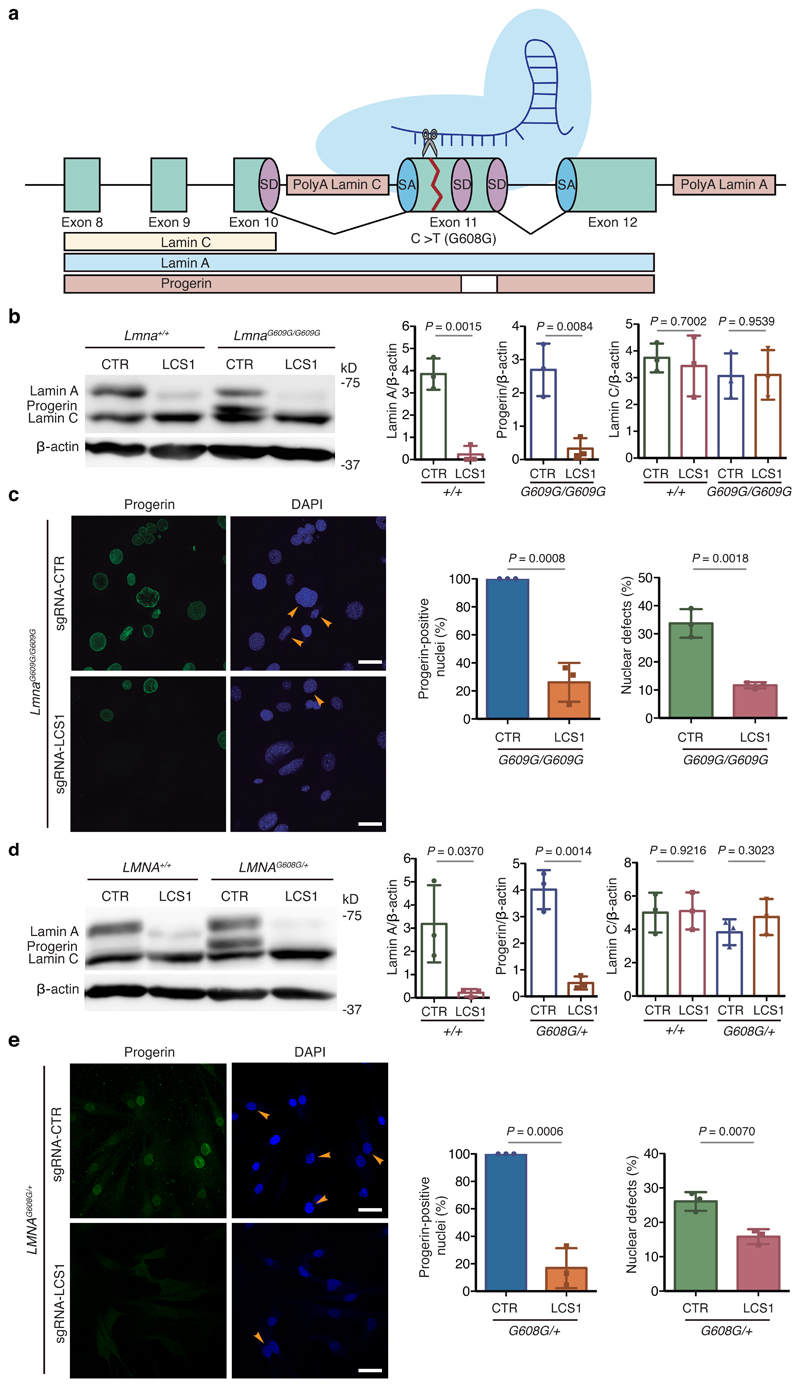
Figure 1. CRISPR/Cas9 testing in HGPS cellular models.
(a) sgRNA-LCS1 directs Cas9 nuclease against exon 11 of LMNA gene upstream of the HGPS mutation, disrupting lamin A and progerin without altering lamin C. (b) Cropped Western blot of lamin A, progerin and lamin C from wild-type and LmnaG609G/G609G mouse embryonic fibroblasts (MEFs) transduced with sgRNA-control or sgRNA-LCS1 (n=3 independent infections and MEF lines; two-tailed Student’s t-test). (c) Immunofluorescence analysis of progerin-positive nuclei and quantification of nuclear alterations by 4′,6-diamidino-2-phenylindole (DAPI) staining (n=3 independent infections and MEF lines; two-tailed Student’s t-test). Arrowheads indicate nuclear aberrations. (d) Cropped Western blot of lamin A, progerin and lamin C from wild-type and LMNAG608G/+ human fibroblasts transduced with sgRNA-control or sgRNA-LCS1 (n=3 independent infections; two-tailed Student’s t-test). (e) Progerin immunofluorescence and analysis of nuclear aberrations by DAPI staining (n=3 independent infections; two-tailed Student’s t-test). Arrowheads indicate blebbings and invaginations. Bar plots represent mean ± SD and individual values are overlaid. Scale bars, 40 μm. Uncropped blots are available as Source Data.