Fig. 4.
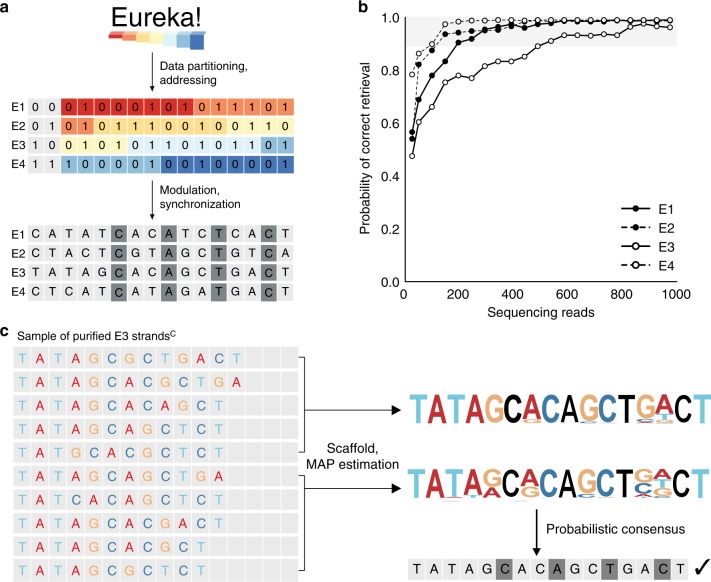
Coded strand architecture for robust information storage. a The message “Eureka!” was encoded and partitioned into four template sequences, E1–E4. Each sequence stores a 2-bit address and 14 bits of data. These bits are mapped to a template sequence of 16 nucleotides, which includes four synchronization nucleotides (dark gray). Synthesis was performed with initiators tethered to beads and sequencing performed on the Illumina platform. b Retrieving information from E1 to E4. Synthesized strandsR were sequenced using the Illumina sequencing-by-synthesis (SBS) platform and purified in silico based on raw length of 32–48 nucleotides (Methods). The decoding accuracy for each sequence is defined as the probability of 100% correct data retrieval for a given number of reads, estimated over 500 decoding trials. Each trial is based on a randomly drawn set of purified strandC variants. A 90% decoding accuracy (gray band) is considered sufficient for robust data retrieval, and this accuracy could be further reinforced by other codec modules. c Decoding of E3. A set of 10 DNA strandsC is decoded as two sets of five strandsC. The decoder uses MAP estimation and a scaffold to determine the probability for each of the four nucleotides at every position. The decoded sequence is a probabilistic consensus of the reconstructed sequences from MAP estimation and successfully retrieves the data stored in E3. Source data for b is provided in the Source Data file package