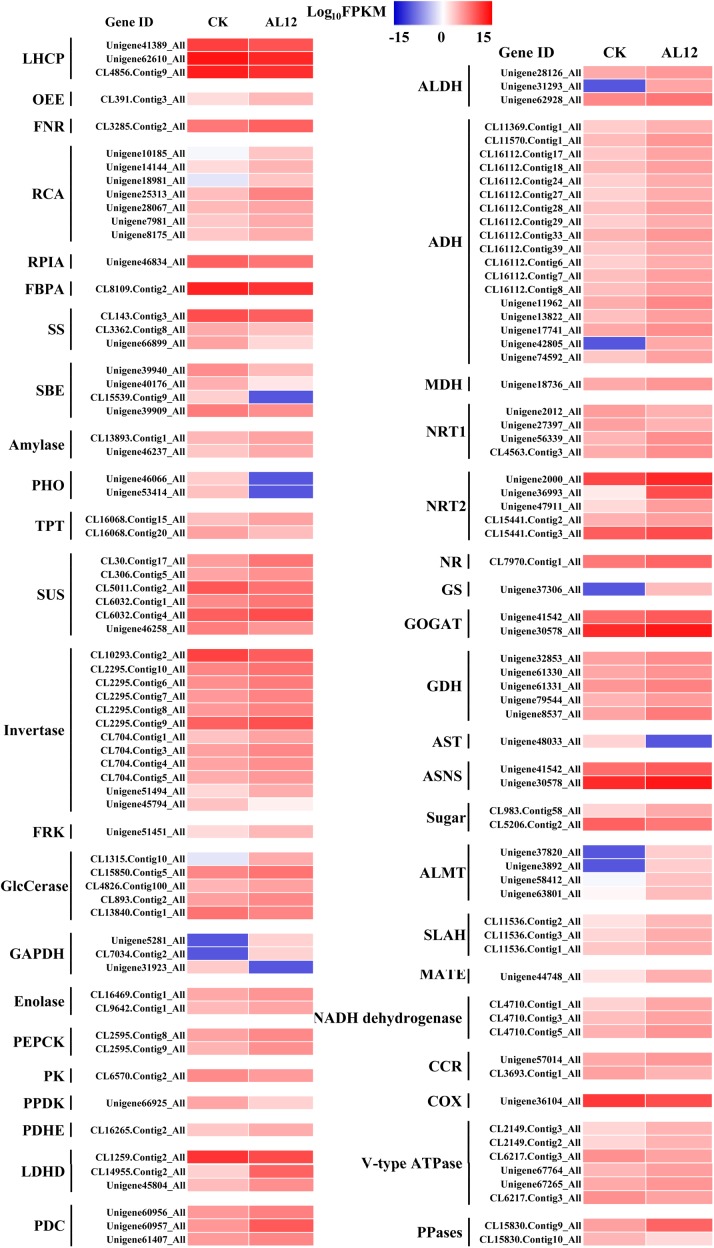
FIGURE 3.
Differentially expression genes involved in photosynthesis, carbon/nitrogen metabolism, and oxidative phosphorylation in shoots of A. lancea after the fungal endophyte Gilmaniella sp. AL12 inoculation. LHCP, light-harvesting chlorophyll a/b-binding protein; OEE, PSII oxygen-evolving enhancer protein; FNR, Ferredoxin-NADP reductase; RCA, ribulose bisphosphate carboxylase/oxygenase activase; RPLA, Ribose 5-phosphate isomerase; FBPA, fructose-bisphosphate aldolase; SS, starch synthase; SBE, 1,4-alpha-glucan branching enzyme; amylase, PHO, starch phosphorylase; TPT, Glucose-6-phosphate/phosphate-translocator; SUS, sucrose synthase; invertase, β-fructosidases; FRK, fructokinase; GlcCerase, Beta-glucosidase-related glycosidases; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PEPCK, phosphoenolpyruvate carboxykinase (ATP); PK, pyruvate kinase; PPDK, Pyruvate orthophosphate dikinase; PDHE, pyruvate dehydrogenase; LDHD, lactate dehydrogenase; PDC, pyruvate decarboxylase; ALDH, aldehyde dehydrogenase (NAD+); ADH, alcohol dehydrogenase; MDH, malate dehydrogenase; NRT1, low-affinity nitrate transporter; NRT2, high-affinity nitrate transporter; NR, nitrate reductase; GS, glutamine synthetase; GOGAT, glutamate synthase; GDH, glutamate dehydrogenase; AST, asparagine transaminase; ASNS, asparagine synthetase; sugar, bidirectional sugar transporter; ALMT, aluminum-activated malate transporter; SLAH, S-type anion channel; MATE, MATE efflux family protein; CCR, cytochrome-c reductase; COX, Cytochrome c oxidase; PPases, Inorganic pyrophosphatase.