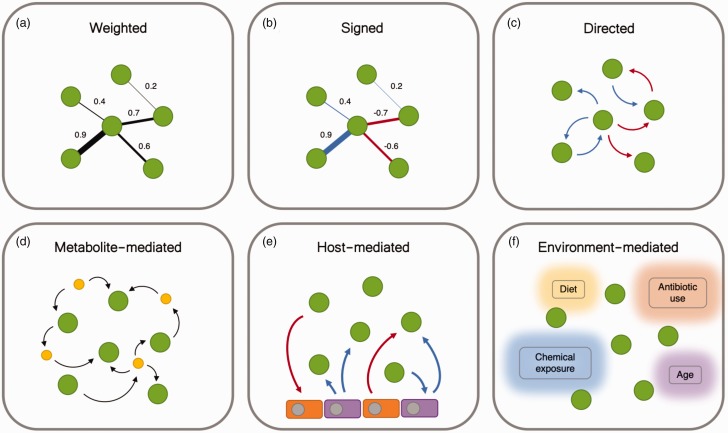
Figure 2.
Network abstractions of the microbial interactome. (a) Weighted networks characterize the strength of an interaction, but do not indicate whether the interaction is mutually positive or negative. Interactions characterized by non-linear relationships take this form. (b) Signed microbial interaction networks denote both the strength and direction of correlations between microbes, but do not indicate a causal relationship. Such networks are typically produced from cross-sectional data. (c) Directed microbial networks characterize source and target of an interaction, indicating a causal relationship. Such networks can be described using the ecological terms in Figure 1, and are typically produced from longitudinal (time-series) data. (d) Interactions between microorganisms are largely mediated by metabolites and macromolecules, which may be consumed or produced as a food source or waste. (e) Host cells play an important role in the microbial ecosystem. Host cells may affect the growth of microbes by secreting metabolites or antibiotics. Microbes break down and produce metabolites and macromolecules like short-chain fatty acids, which act as an energy source, and promote the differentiation of host cells in turn. (f) Environment-mediated microbial interaction networks contain context-dependent edges. These variables may conditionally alter the topology and dynamics of the microbial interaction networks.