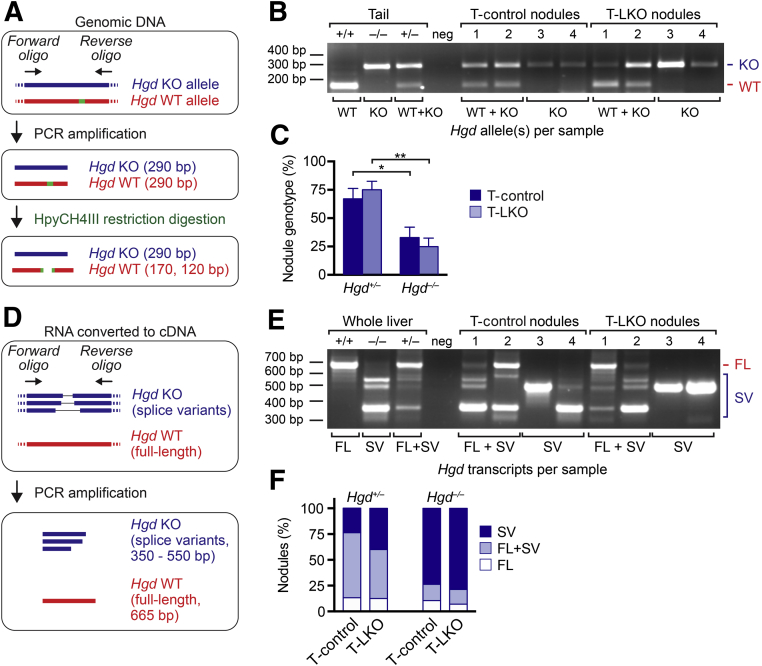
Figure 6.
The Hgd wild-type (WT) allele was lost by similar mechanisms in equal frequencies by regenerating nodules from Tyrosinemia-control (T-control) and Tyrosinemia–liver-specific E2f7/E2f8 double knockout (T-LKO) livers. A–C: Genomic DNA analysis for each nodule. Strategy for detecting Hgd WT and knockout (KO) alleles in genomic DNA by PCR. Genomic DNA was amplified by PCR and was digested with restriction enzyme HpyCH4III (A). Tail tissue DNA isolated from Hgd+/+, Hgd–/–, and Hgd+/– mice illustrate the WT (170 bp) and KO (290 bp) bands. The 120-bp WT band is not shown. Neg is a negative control that did not contain template DNA. Representative nodules from T-control and T-LKO livers depict WT + KO alleles (nodules 1 and 2) and KO alleles (nodules 3 and 4; B). Nearly 25% of regenerating nodules from T-control and T-LKO livers lost the Hgd WT allele and are effectively Hgd–/– (C). D–F: Transcriptomic analysis of Hgd transcripts in each nodule. Strategy for detecting full-length (FL) transcript from the Hgd WT allele or splice variants (SVs) from the Hgd KO allele (D). Reverse transcription-PCR with the use of RNA from whole livers isolated from Hgd+/+, Hgd–/–, and Hgd+/– mice illustrate FL and SV banding patterns. Neg is a negative control that did not contain template cDNA. Representative nodules from T-control and T-LKO livers depict FL + SV Hgd transcripts (nodules 1 and 2) and SV Hgd transcripts (nodules 3 and 4; E). The percentage of Hgd transcripts is indicated for nodules genotyped as Hgd+/– and Hgd–/–. Hgd transcripts were expressed similarly in nodules from T-control and T-LKO mice, and, consistent with loss of the WT Hgd allele, up to 80% of Hgd–/– nodules contained Hgd SVs only (F). Data are expressed as means ± SEM (C) or means only (F). n = 57 nodules from 4 T-control mice; n = 54 nodules from 4 T-LKO mice. ∗P < 0.05, ∗∗P < 0.01.