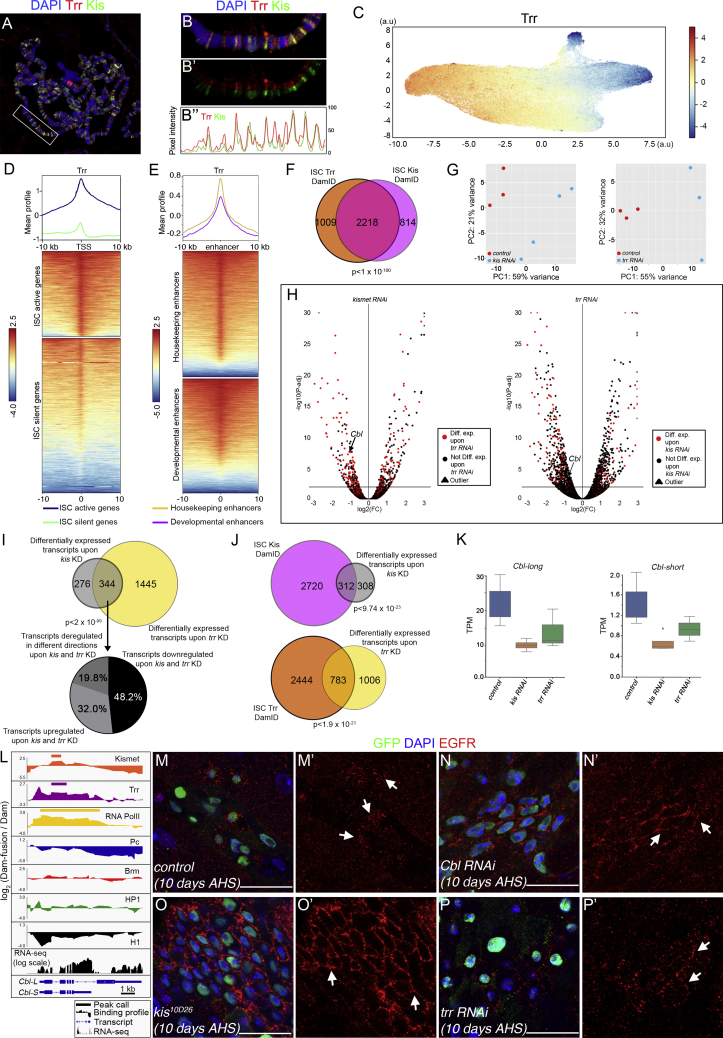
Figure 6.
Genome-wide Mapping of Trr DNA Binding Sites and Genes Regulated by Kismet and Trr
(A) Kismet and Trr localization on polytene chromosomes of the salivary gland.
(B–B″) Magnification of the chromosome highlighted in (A). Fluorescent intensity in (B″).
(C) Trr binding in the ISC clustered using UMAP based on 7 DamID fusion proteins (STAR Methods and Figure 4C).
(D) Mean position and metaplot of Trr in ISCs plotted relative to the TSS for genes according to their activity based on RNA Pol II occupancy shows its enrichment over the TSS of active genes.
(E) Mean position and metaplot of Trr in ISC over previously defined “developmental” or “housekeeping” enhancers in S2 cells from Zabidi et al., 2015.
(F) Overlap between genes with peaks of Kismet versus Trr.
(G) Principal-component analysis of RNA-seq.
(H) Differentially expressed genes; red points highlight common genes.
(I) Upper: overlap between genes with RNAs deregulated upon kis and trr knockdown in the ISCs. Lower: proportion of RNAs altered in kis and trr knockdown.
(J) Upper: genes with peaks of Kismet and deregulated after kis knockdown in the ISCs. Lower: genes with peaks of Trr and deregulated after trr KD in the ISCs.
(K) RNA-seq data showing downregulation of the 2 Cbl isoforms upon either kis RNAi and trr RNAi in the ISC.
(L) Alignement at the Cbl locus of wild-type RNA-seq, Kismet, Trr, RNA Pol II, Pc, Brm, HP1, H1 binding profiles and peaks as determined by DamID-seq in ISCs.
(M–P′) clone of wild-type (M and M′), Cbl RNAi (N and N′), kis10D26 (O and O′) and trr RNAi (P and P′), 10 days AHS. Arrows show EGFR-positive cells. Scale bars, 20 μm.