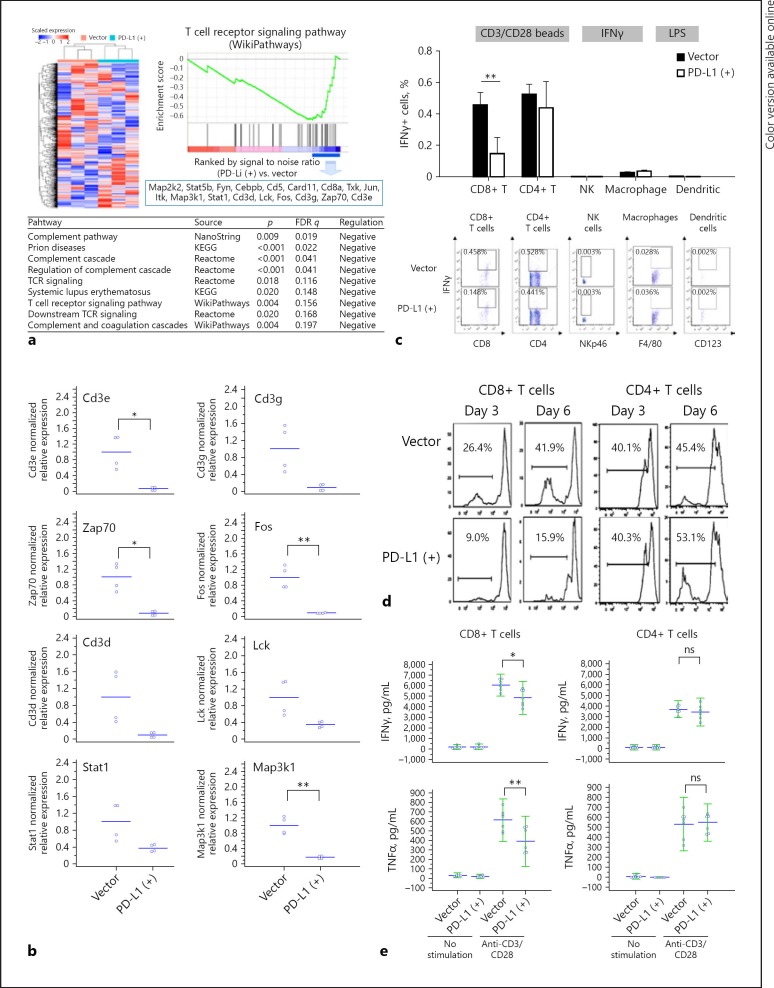
Fig. 4.
Immune microenvironment in HCC of the orthotopic mouse model. a Genes related to immune cells in PD-L1-expressing and parental tumors were analyzed by gene ontology enrichment analysis and NanoString nCounter® PanCancer Immune Profiling Panel. The heatmap and dendrogram showing distinct gene patterns were generated using GAP software with Pearson correlation and complete linkage. b QRT-PCR revealed downregulation of genes related to CD8+ T-cell signaling. Downregulated or upregulated genes were selected under the criteria (1) log fold change ≤0.67 (fold change < 0.2) or > 0.7 (fold change > 5), (2) p value < 0.1. c Mouse splenocytes were co-cultured with the BNA-MEA liver cancer cells (with or without PD-L1 expression). The expression of interferon-γ (IFNγ) in T cells, NK cells, and macrophage/dendritic cells was measured by intracellular staining and flow cytometry after the splenocytes were stimulated with CD3/CD28 beads (for T cells), IFNγ (for NK cells), or lipopolysaccharides (for macrophage/dendritic cells). IFNγ expression in CD8+ T cells was significantly suppressed by PD-L1 expression in liver cancer cells, while those in CD4+ T cells, NK cells, and macrophage/dendritic cells were not affected significantly. d, e CD8+ or CD4+ T cells isolated from mouse splenocytes were co-cultured with BNL-MEA liver cancer cells with or without PD-L1 expression. In CD8+ T cells proliferation (d) and release of IFNγ or tumor necrosis factor-α (TNFα) (e) after stimulation were significantly suppressed when co-cultured with tumor cells expressing PD-L1. By contrast, in CD4+ T cells neither proliferation nor cytokine release were affected significantly. Values are presented as means ± SD (n = 5 in each group). Statistical significance was set at * p < 0.05, ** p < 0.01, statistically significant (one-way ANOVA).