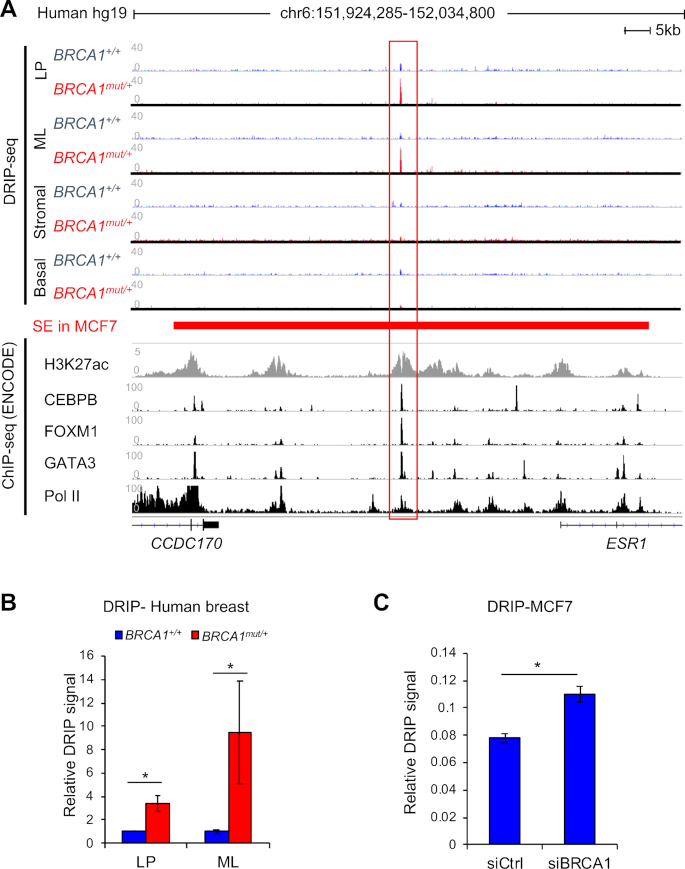
Figure 1.
BRCA1-associated R-Loop accumulation at a non-coding region upstream of ESR1 locus. (A) Alignment of DRIP-seq profiles in primary human breast cells with functional annotations of other published genomic data in MCF7 cells through ENCODE project. DRIP-seq was done in four sorted populations: luminal progenitor (LP), mature luminal (ML), stromal cells, and basal epithelial cells. Each track is an overlay of four individual non-carriers (BRCA1+/+) or four BRCA1 mutation carriers (BRCA1mut/+) indicated by different colors. ENCODE ChIP-seq profiles of various transcription factors are labeled for each track. The super-enhancer region defined by a published study in MCF7 cells is represented by a red bar (40). All sequence data are aligned to human reference genome, hg19. (B) Validation of DRIP-seq result using locus-specific DRIP. *P < 0.05 by Student's t-test. Error bar: s.e.m. (C) Locus-specific DRIP in control and BRCA1 knockdown MCF7 cells. Experiments were performed three times. *P < 0.05 by Student's t-test. Error bar: s.e.m.