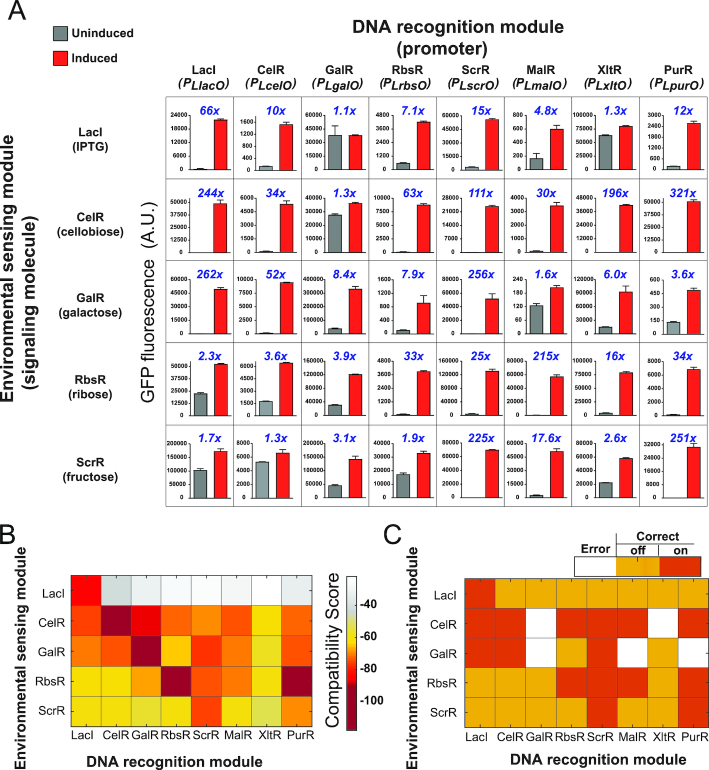
Figure 2.
Characterization of LacI family hybrid repressors via experimental design and coevolutionary models. (A) Experimental characterization of hybrid repressors. We assessed the ability of hybrid repressors in gene expression regulation and allosteric response, by using a transcriptional reporter assay in a strain of E. coli with GFP as a reporter of transcription activities. A number of 40 repressors (including 35 hybrids) were characterized with this assay. Results shown in each column and each row are from repressors with the same DRM and ESM, respectively. Repressors with the same ESM were exposed to the same signaling molecule as indicated in the bracket on the left of each row. Repressors with the same DRM were used to control GFP expression driven by the same promoter (shown for each column). Data points in bar graphs represent GFP fluorescence in cells that were uninduced (grey) and induced (red). The blue number above each plot represents the corresponding fold-change of GFP induction. All data points represent mean ± S.D. of three biological replicates. (B) Heatmap of compatibility scores, C(S), predicted from coevolutionary information for 40 hybrid repressors. The hybrid repressors with the same ESM are presented in rows, while the repressors with the same DRM are shown in columns. The more negative score is shown in darker color, indicating more favorable compatibility. (C) Agreement between experimental observations and coevolutionary compatibility predictions. Orange and red cells indicate successful prediction of negative or positive for hybrid functionality (induction threshold at 20), respectively. White cells indicate disagreement between computational predictions and experimental results.