Figure 4.
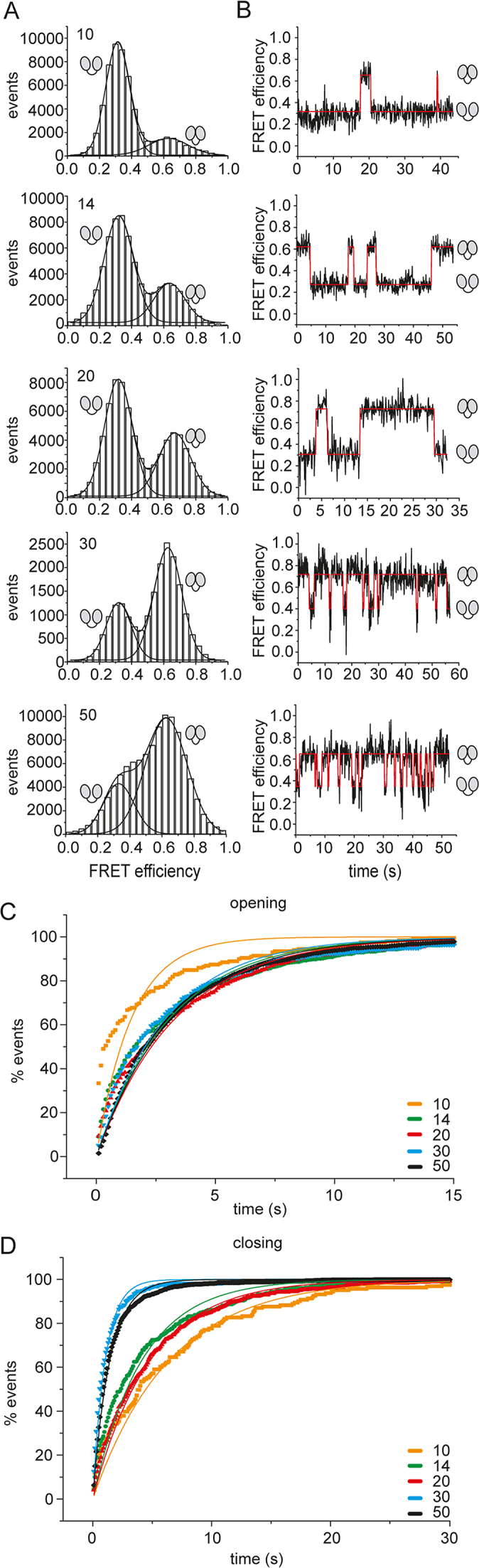
Effect of single-stranded RNAs of different lengths on the eIF4A conformational cycle. (A) FRET histograms for eIF4A in the presence of eIF4B, eIF4G, and the 10mer, 14mer, 20mer, 30mer, 40mer, or 50mer single-stranded RNAs. Experiments were performed with biotinylated, donor/acceptor-labeled, surface-immobilized eIF4A in the presence of 10 μM eIF4B and eIF4G, and 15 μM of the 10mer, 14mer, 20mer, 30mer, 40mer, or 50mer RNA, and 3 mM ATP in 50 mM Tris/HCl, pH 7.5, 80 mM KCl, 2.5 mM MgCl2, 1 mM DTT and 1% glycerol at 25°C. (B) Representative FRET time traces. (C) Normalized cumulative dwell time histograms for opening and single-exponential fits. See Supplementary Figure S2A for analyses with single-exponential functions and the respective residuals, Supplementary Figure S3A for analyses with double-exponential functions and residuals, and Supplementary Table S1 for corrected R2-values. (D) Normalized cumulative dwell time histograms for closing and single-exponential fits. See Supplementary Figure S2B for single-exponential fits and respective residuals, Supplementary Figure S3B for double-exponential fits and residuals, and Supplementary Table S1 for corrected R2-values. The cartoons indicate the low-FRET (half-)open conformation and the high-FRET closed state of eIF4A.
