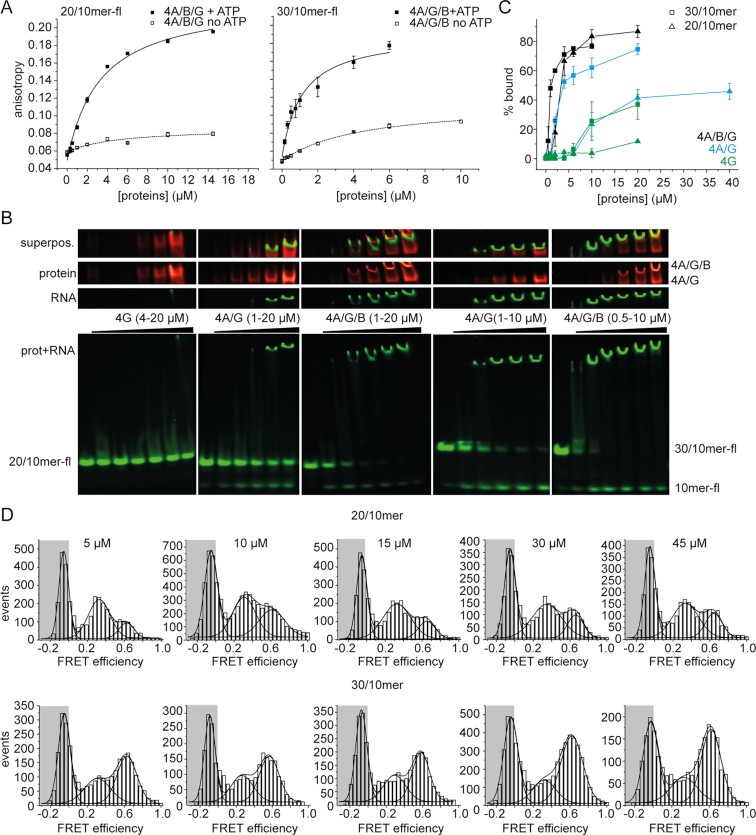
Figure 6.
Interaction of eIF4A with 20/10mer and 30/10mer RNA. (A) Anisotropy titrations of fluorescein-labeled 20/10mer and 30/10mer RNAs with eIF4A/B/G in the absence (open squares) and presence of 5 mM ATP (filled squares). Titrations were performed in 30 mM HEPES/KOH, pH 7.4, 100 mM KOAc, 3 mM Mg(OAc)2, and 2 mM DTT at 25°C. Error bars depict the standard error of the mean from two independent experiments. (B) Electrophoretic mobility shift assay of 100 nM fluorescein-labeled 20/10- and 30/10mer with increasing concentrations of eIF4G, eIF4A/G and eIF4A/B/G in 30 mM HEPES/KOH, pH 7.4, 100 mM KOAc, 3 mM Mg(OAc)2, 2 mM DTT in the presence of 5 mM ATP and 0.4 U/μl of RNase inhibitor, incubated for 10 min at 25°C. To regenerate ATP, 23 μg/ml pyruvate kinase and 1 mM phoshoenolpyruvate were added. 1% of eIF4G is labeled with Alexa647 (red), the 10mer of the RNA substrate is labeled with fluorescein (fl; green). A representative gel from two independent EMSAs is shown. Note that these epxeriments were performed with an excess of translation initiation factors, such that under saturation all of the RNA is protein-bound, but only a small fraction of the translation factors is bound to RNA. See Supplementary Figure S8 for control experiments with labeled eIF4A and labeled eIF4B. (C) Quantification of RNA-bound complexes of eIF4G, eIF4A/G, and eIF4A/B/G from B. Error bars depict either the error of the mean from two independent experiments (20/10mer, eIF4G; 20/10mer, eIF4A, eIF4G, eIF4B) or the standard deviation from at least three independent experiments (all other experiments). (D) smFRET experiments in presence of different concentrations of 20/10mer and 30/10mer RNAs. 100 pM biotinylated eIF4A_Q186C/G370C, labeled with Alexa488-maleimide (A488, donor) and Alexa546-maleimide (A546, acceptor), in 50 mM Tris/HCl, pH 7.5, 80 mM KCl, 2.5 mM MgCl2, 1 mM DTT, and 1% glycerol in the presence of 3 mM ATP, 10 μM eIF4B and eIF4G, and 5, 10, 15, 30, and 45 μM RNA at 25°C.