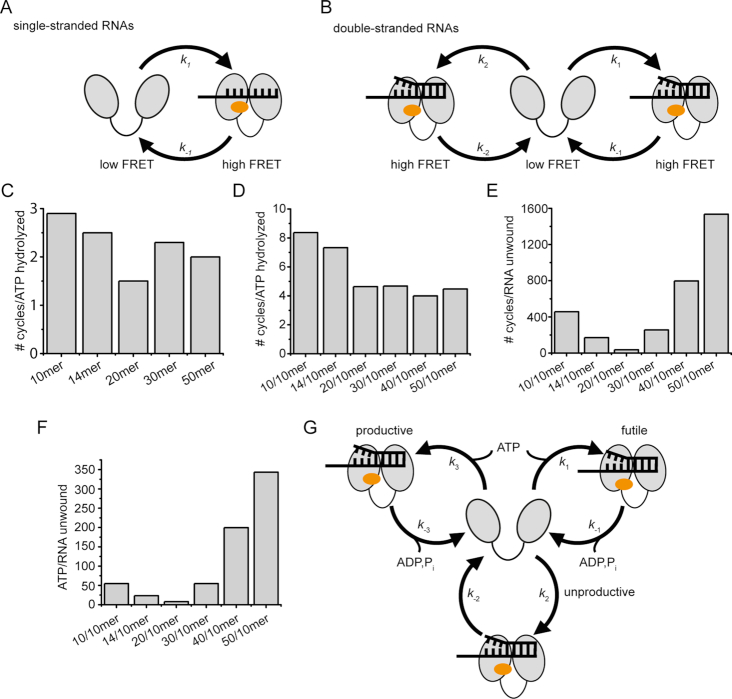
Figure 7.
Kinetic models for conformational cycling of eIF4A, and comparison of conformational dynamics, ATP hydrolysis, and RNA unwinding. (A) Kinetic model for eIF4A conformational changes in the presence of single-stranded RNAs and RNAs with long 5′-single-stranded regions. (B) Kinetic model for conformational cycling in the presence of RNAs with no or short 5′-single-stranded regions. (C) Coupling of conformational cycling to ATP hydrolysis in the presence of single-stranded RNAs. The number of conformational cycles per ATP hydrolyzed is independent of the length of the RNA. (D) Number of conformational cycles per ATP hydrolyzed in the presence of double-stranded RNAs. (E) Number of conformational cycles per RNA unwound. In the presence of the 20/10mer RNA, eIF4A undergoes the minimal number of approx. 37 conformational cycles per RNA unwound. (F) Coupling of ATP hydrolysis to RNA unwinding. In the presence of the 20/10mer RNA, eIF4A hydrolyzes the minimal number of 8 ATP per RNA unwound. (G) Model linking eIF4A conformational cycling with ATP hydrolysis and RNA unwinding. eIF4A can undergo futile cycles (ATP hydrolysis, but no RNA unwinding), unproductive cycles (no ATP hydrolysis, no RNA unwinding), and productive cycles (ATP-dependent RNA unwinding). In productive cycles the first strand of an RNA duplex dissociates from eIF4A closed state, prior to ATP hydrolysis. In futile cycles the duplex dissociates from eIF4A without unwinding, and ATP is then hydrolyzed. In unproductive cycles, eIF4A undergoes conformational changes without ATP hydrolysis and unwinding. The kinetic competition of the individual cycles determines the coupling of conformational cycling, ATP hydrolysis, and RNA unwinding. The cartoons reflect the conformation of eIF4A in the presence of the translation initiation factors eIF4B and eIF4G. Note that for simplicity the closed states in the different conformational cycles in panels B and G are depicted identically, but reflect different physical states with different functional properties.