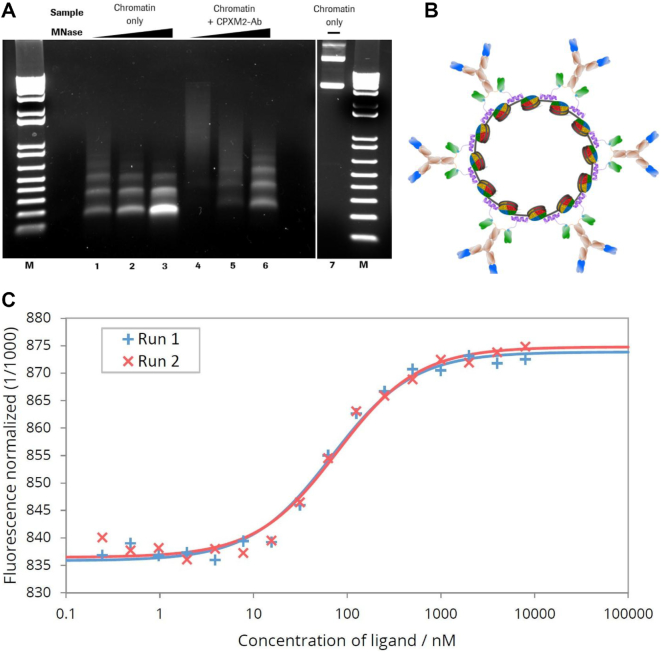
Figure 1.
MNase digestion of antibody–chromatin complexes and antibody-complexation with plasmid-chromatin. (A) Agarose gel electrophoresis of chromatin without (lane 1–3) and in presence of biotin-CPXM2 ∼ anti biotin 2+2 bsAb constructs (lane 4–6) after partial DNA hydrolysis by MNase with increasing incubation time (20s, 80s and 270s); Chromatin without MNase treatment (lane 7) is shown as control. Mononucleosomal DNA bands (147bp) indicate complete digestion in contrast to higher molecular weight bands. In presence of biotin-CPXM2 ∼ anti biotin 2+2 bsAb constructs, chromatin is more nuclease resistant as the 147 bp DNA band only occurs at late time-points of Nuclease treatment in comparison to the chromatin only sample. (B) Scheme of antibody–chromatin complexes with plasmid DNA reconstituted into nucleosomes and associated antibody-peptide constructs. Variable regions against cell surface antigen (blue) faces outwards and anti biotin scFv (green) is bound at biotin-CPXM2 peptide (purple) associated at the DNA backbone. (C) MST runs for Chromatin + biotin-CPXM2 ∼ anti biotin 2+2 bsAb interaction. Ligand concentration refers to biotin–CPXM2 peptide (twice as much as the respective anti biotin 2+2 bsAb concentration). Exp 1 (blue) and Exp 2 (red) are independent experiments of the same construct with the respective curve fit for KD determination.