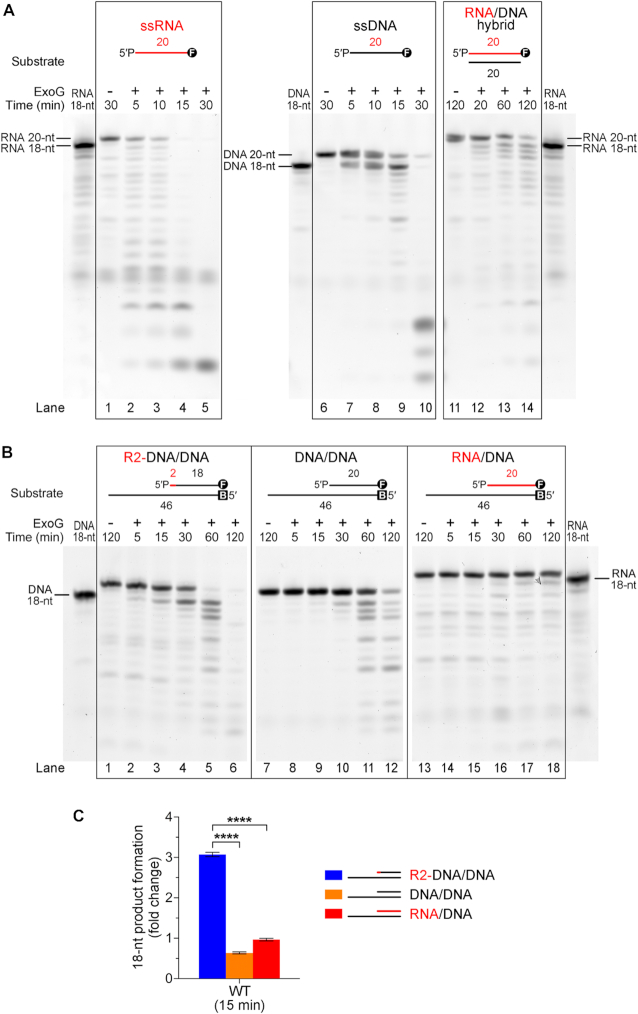
Figure 1.
ExoG preferentially removes 5′-end RNA dinucleotide in an RNA–DNA chimeric duplex. (A) Time-course nuclease activity assays of wild-type ExoG (50 nM) degrading the 3′-end FAM-labeled 20-nt ssRNA, ssDNA and 20-bp RNA/DNA hybrid duplex substrates (100 nM). The 18-nt RNA and DNA markers are shown at the sides of the gel. (B) Time-course nuclease activity assays of wild-type ExoG (25 nM) degrading R2-DNA/DNA, DNA/DNA and RNA/DNA substrates (100 nM). Markers for 18-nt DNA and RNA are displayed on either side of the gels. DNA substrates are labeled with FAM (marked as circled F) at 3′ end for detection, and/or biotin at 5′ end (marked as B) for capping with NeutrAvidin in the reaction condition to block the activity of ExoG. (C) Quantification of the 18-nt product generated from R2-DNA/DNA, DNA/DNA and RNA/DNA substrates by wild-type ExoG at 15 min. Error bars represent the standard errors from at least four replicates of the experiment. Statistical significance (P values) was determined by an unpaired two-tailed Student's t-test, with **** representing P < 0.0001.