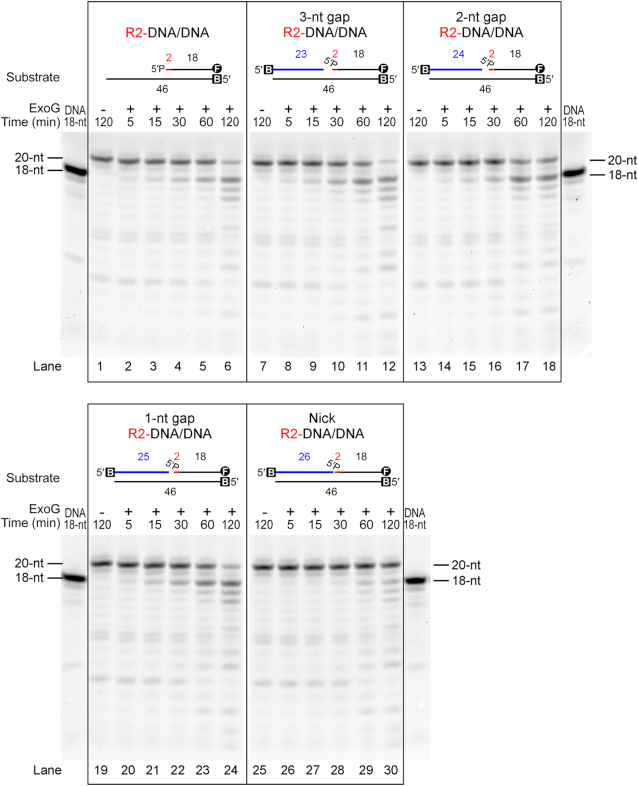
Figure 2.
ExoG removes the 5′-end RNA dinucleotide in gap DNA duplexes. Time-course nuclease activity assays show that wild-type ExoG (25 nM) degraded R2-DNA/DNA (lanes 1–6) and R2-DNA/DNA annealed with additional 3′-end upstream DNA strands (displayed in blue) to produce DNA substrates with 3-, 2- or 1-nt gap (lanes 7–24). ExoG had limited activity in degrading the nick duplex substrates (100 nM) (lanes 25–30). Markers for 18-nt DNA is displayed on both side of the gels. DNA substrates are labeled with FAM (marked as circled F) at 3′ end for detection, and/or biotin at 5′ end (marked as B) for capping with NeutrAvidin in the reaction condition to block the activity of ExoG.