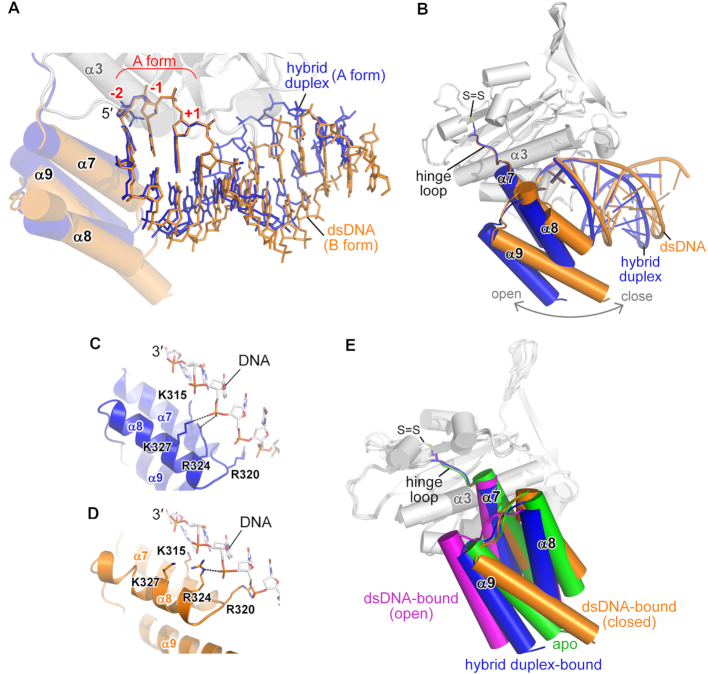
Figure 5.
A flexible C-terminal Wing domain allows ExoG to bind diverse substrates. (A, B) Superimposition of the ExoG–RNA/DNA (in blue) and ExoG–DNA (in yellow) complexes, focusing on the bound substrates (A) and C-terminal Wing domain (B). In A, the well-aligned –2 to +1 nucleotides that adopt the A-form conformation are labeled in red. (C, D) Interaction between the non-scissile DNA strands and the Wing domains in the ExoG–RNA/DNA (blue) and ExoG–DNA (yellow) complexes. (E) Superimposition of available structures of ExoG. The Wing domains of the respective structures are colored as follows: ExoG–RNA/DNA structure (blue; pdb ID: 5ZKJ, this study); ExoG–DNA structure (yellow; pdb ID: 5ZKI, this study); ExoG–DNA structure (magenta; pdb ID: 5T5C) (30); and ExoG apo-form structure (green; pdb ID: 5T40) (30). The disulfide bond (labeled as S = S) formed between C294 and C299 in all structures is shown as sticks. In all structures, the enzyme core domain (in gray) was used for protein secondary structure superimposition.