Figure 1.
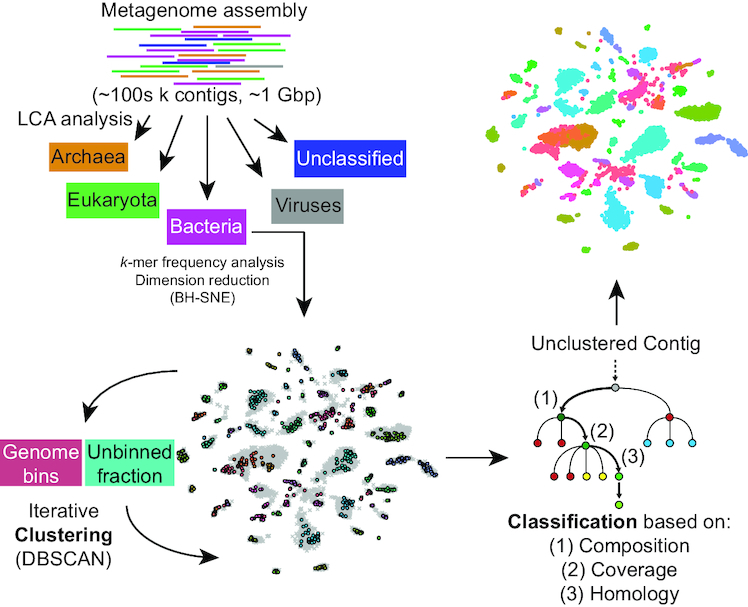
Autometa binning workflow. Autometa separates contigs from a de novo metagenome assembly into kingdom-level bins based on sequence homology, iteratively clusters kingdom-specific (Bacterial or Archaeal) contigs, and then (optionally) classifies any remaining unclustered contigs to bins using a decision tree classifier.
