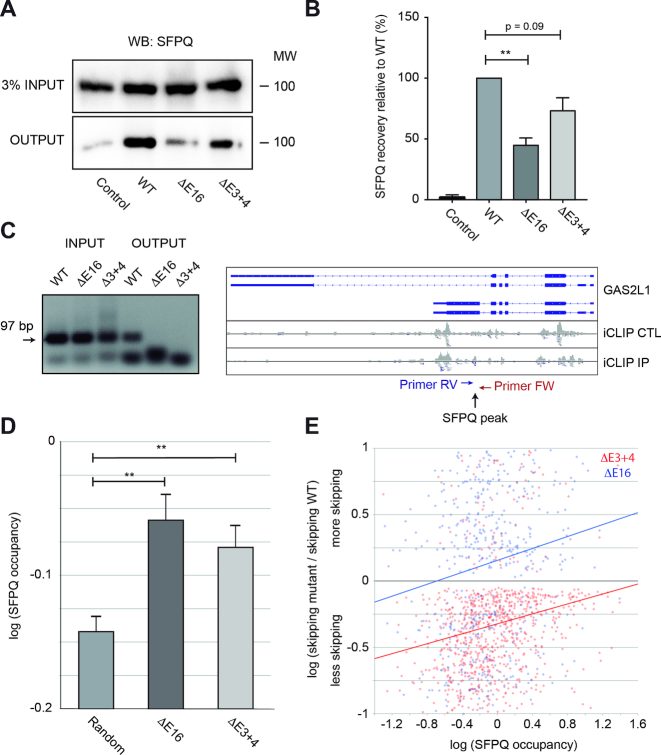
Figure 4.
Dido mutation reduces SFPQ binding to RNA. (A) SFPQ binding to RNA was assayed by PAR-CLIP. While robust binding was observed in WT MEF, SFPQ signal was partially lost in the Dido mutants. Inputs showed comparable SFPQ expression levels in all samples. (B) Quantitation of three independent experiments showed significant loss of SFPQ binding in E16 and a similar tendency in E3+4. Error bars represent standard deviation (**P < 0.01). (C) iCLIP followed by RT-PCR confirmed lack of SFPQ binding to the GAS2L1 target, in which exon skipping increased. Positions of SFPQ binding and RT-PCR primers are indicated (right). Next, SFPQ binding to the 5′ end of exons was quantified from iCLIP data (iCLIP IP) and corrected for expression (iCLIP CTL). (D) Average SFPQ binding to alternatively used exons was significantly higher than SFPQ binding in a random control group. Error bars represent standard error of the mean (**P < 0.01). (E) SFPQ binding levels of individual exons in WT cells correlate positively with increased skipping in E16 and E3+4, indicating high SFPQ dependency of affected exons.