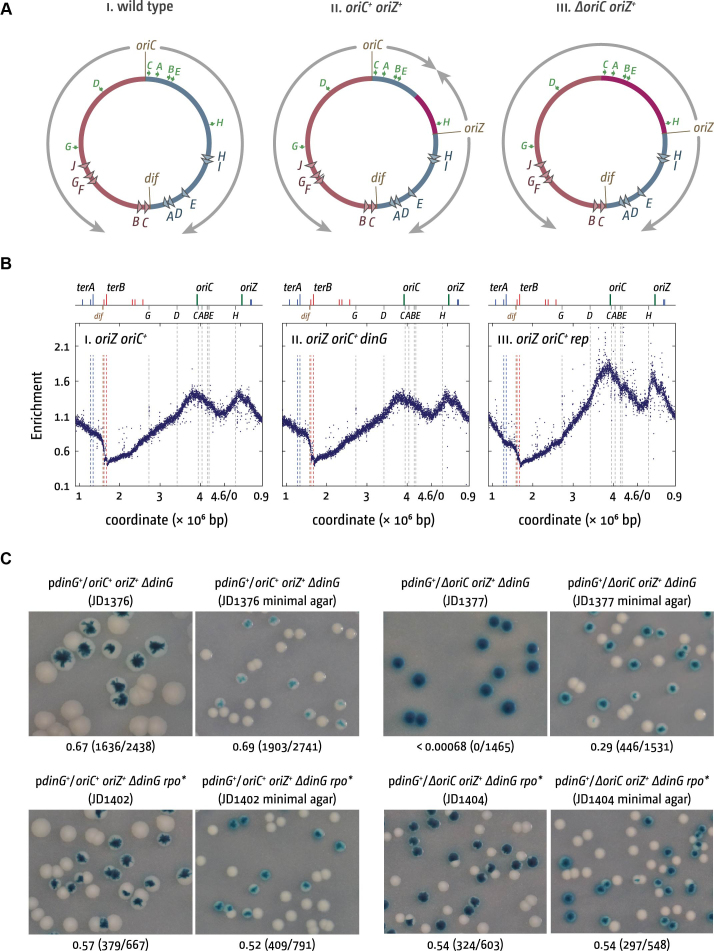
Figure 6.
Maintenance of chromosome replication and cell viability of oriC+ oriZ+ and ΔoriC oriZ+ cells in the absence of DinG helicase. (A) Schematic representation of the replichore arrangement of one or two replication origins in E. coli. The origins oriC and oriZ as well as the dif chromosome dimer resolution site are highlighted. Replichores and replication directionality are indicated by grey arrows. ter sites are indicated by triangles and identified by their corresponding letter (‘A’ indicates the terA site). Green arrows represent location and direction of transcription of the 7 rrn operons A–E, G and H. Chromosomal sections in which the direction of DNA replication is artificially inverted because of the presence of an ectopic replication origin are shown in purple. (B) Replication profiles of oriC+ oriZ+ strains in the absence of either DinG or Rep helicases. The replication profiles are generated by plotting the number of sequence reads (normalized against reads for a stationary phase wild type control) against their chromosomal location. The schematic representation of the E. coli chromosome above each panel shows the positions of the two origins, oriC and oriZ, and ter sites (above) as well as the dif chromosome dimer resolution site and rrn operons A–E, G and H (below). The strains used were RCe504 (oriC+ oriZ+) and JD1277 (oriC+ oriZ+ ΔdinG). Data for JD1141 (oriC+ oriZ+ Δrep) were re-plotted from Dimude et al. (59). All three constructs were sequenced in parallel. (C) The plate photographs shown are of synthetic lethality assays, as described in Supplementary Methods. The relevant genotype of the construct used is shown above each photograph, with the strain number in parentheses. The fraction of white colonies is shown below, with the number of white colonies/total colonies analysed in parentheses. The plasmid used was pJD001 (dinG+) (see Supplementary Information).