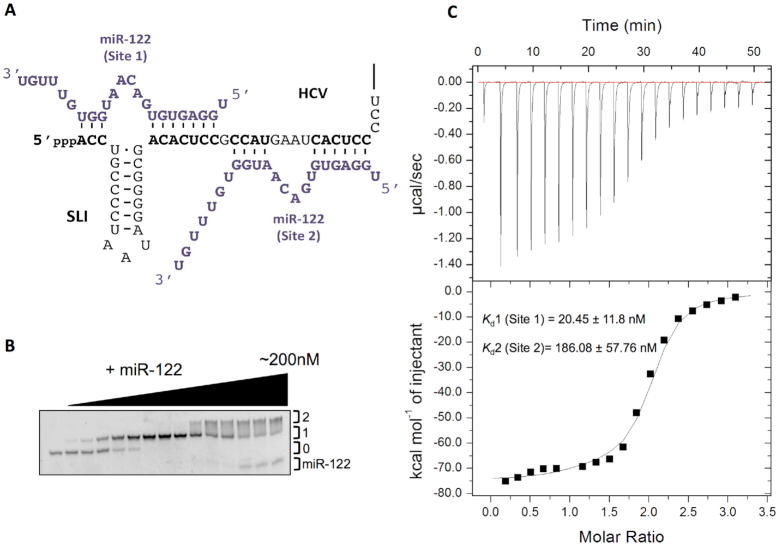
Figure 1.
miR-122 binds to the HCV 5′ terminus with a 2:1 stoichiometry. (A) Model of the interaction of miR-122 (purple) and nts 1–42 of the HCV genome (black). (B) Non-denaturing gel electrophoretic mobility shift assay (EMSA) of 1–42 nt HCV RNA and increasing amounts of miR-122. (C) Thermogram and resulting binding curve of the titration of 1–42 nt HCV RNA with miR-122. Affinities of each binding site, Kd1 (Site 1) and Kd2 (Site 2), are indicated. All data are representative of at least three independent replicates and Kd values are an average of three measurements (±) error propagated from individual fits.