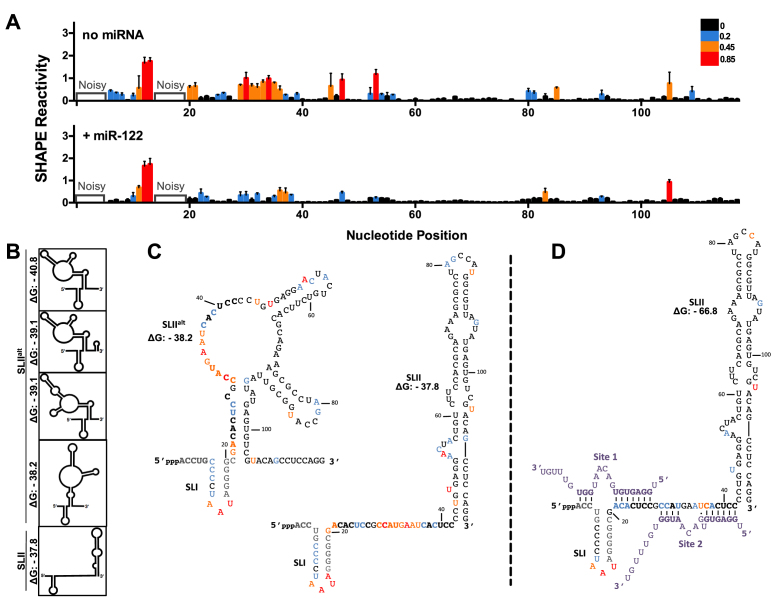
Figure 2.
miR-122 binding suppresses an alternative structure (SLIIalt) and promotes functional folding of SLII. (A) Normalized SHAPE reactivities of HCV RNA (nts 1–117) with no miRNA (top) or in the presence of miR-122 (bottom). Nucleotides 1–5 and 14–19 were omitted due to high background reactivity. Nucleotides with high (≥0.85, red), intermediate (between 0.4 and 0.85, orange), low (between 0.2 and 0.4, blue) and very low (≤0.2, black) SHAPE reactivity are indicated. (B) Predictions of the lowest free energy structures formed by the first 117 nts of HCV 5′ UTR and their predicted free energies (ΔG, kcal/mol). (C) SHAPE data from (A) superimposed onto two of the predicted structures of HCV RNA alone or (D) HCV RNA + miR-122, where the predicted structure and free energy values are indicated. The SHAPE data represents the results of six independent replicates and error bars indicate the standard error of the mean (SEM).