Figure 5.
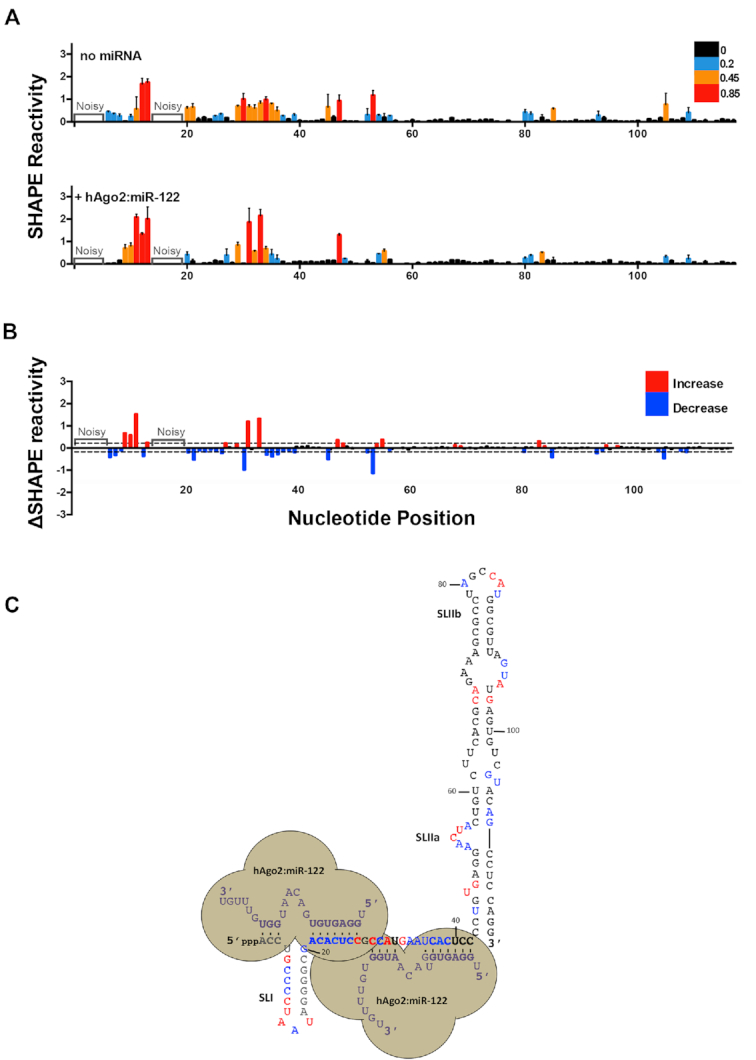
hAgo2:miR-122 complexes alter the secondary structure of the 5′ terminus and SLII of the HCV genome. (A) Normalized SHAPE reactivities of 1–117 nt HCV RNA in the absence (top) or presence of hAgo2:miR-122 (bottom). Nucleotides 1–5 and 14–19 were omitted due to high background reactivity. (B) Difference plot showing changes in SHAPE reactivity upon miR-122 binding, ΔSHAPE = hAgo2:miR-122:HCV RNA reactivity – HCV RNA (no miRNA) reactivity. Significant increases in reactivity (above baseline, red) and decreases in reactivity (below baseline, blue) are indicated. (C) Changes in reactivity upon hAgo2:miR-122 binding are superimposed onto the hAgo2:miR-122:HCV RNA secondary structure model (coloured as in (B)). The data represents the results of four independent replicates and error bars represent the standard error of the mean (SEM).
