Table 2.
Conservation of the 25% strongest human origins according to their association with TSSs and CGIs: percentage of origins overlapping conserved genomic segments, and percentage of functionally conserved origins
| Human–chicken | Human–mouse | |||
|---|---|---|---|---|
| Origin category | % Overlapping a CGS | % Functionally conserved | Overlapping a CGS | % Functionally conserved |
| All | 48.6% 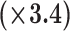
|
19.8% 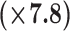
|
82.5% 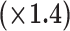
|
32.5% 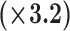
|
| CGI[−] TSS[−] | 33.8% | 11.6% | 75.2% | 22.5% |
| CGI[+] TSS[−] | 63.1% | 29.9% | 82.7% | 36.5% |
| CGI[−] TSS[+] | 58.4% | 21.0% | 92.2% | 37.0% |
| CGI[+] TSS[+] | 70.7% | 33.4% | 95.1% | 52.4% |
| Dormant origins | 11.4% 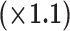
|
- | 63.1% 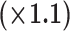
|
- |
Dormant origins are defined as HeLa MCM7 Chip-Seq peaks (23) that do not overlap HeLa origins detected by SNS-seq. Functional conservation could not be assessed for dormant origins since we only have data on the human HeLa cell line (and not for chicken nor mouse).
