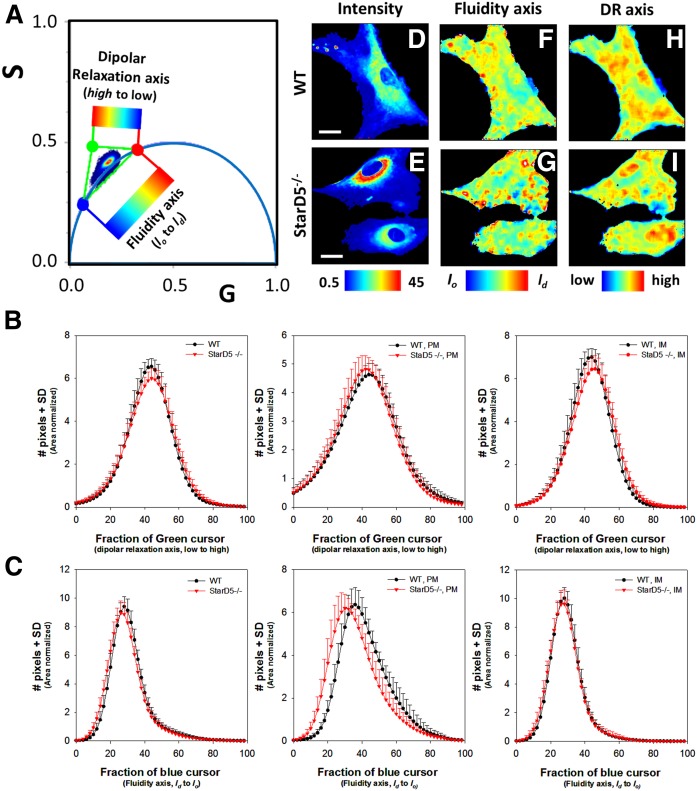
Fig. 7.
Phasor-FLIM of LAURDAN fluorescence in WT and StarD5−/− macrophages for green-channel. A: Three cursors fraction histogram analysis on the phasor distribution for the green channel. The analysis calculates an area normalized number of pixels in the phasor distribution between red-green and red-blue cursors. We define the trajectory between red-green points as the dipolar relaxation axis and from red-blue points as the fluidity axis. The images can be colored by using the scale shown. The values for the G and S coordinates were calculated as detailed in the Materials and Methods section. B: Histograms of the pixel fraction distribution along the red-green trajectory for total membrane, PM, and internal membrane (IM), respectively, from left to right. C: Histogram of the pixel fraction distribution along the red-blue trajectory for total membrane, PM, and internal membrane (IM), respectively. D, E: Representative intensity fluorescence images of WT (D) and StarD5−/− (E) macrophages. The bar at the bottom represents the intensity scale in counts. F, G: Pseudocolor images produced by the cursor selection between red-green cursors (fluidity axis). The bar at the bottom represents the color scale used for the pseudo-coloring between the lo/ld trajectory. H, I: Pseudocolor images produced by the cursor selection between red-blue cursors (dipolar relaxation axis). The bar at the bottom represents the color scale used for the pseudo-coloring between low and high dipolar relaxation trajectory. The separation of plasma and internal membranes was done using masks. In the trajectory analysis, we used more than 20 cells by group. Bars represent 20 μm.