Fig. 3.
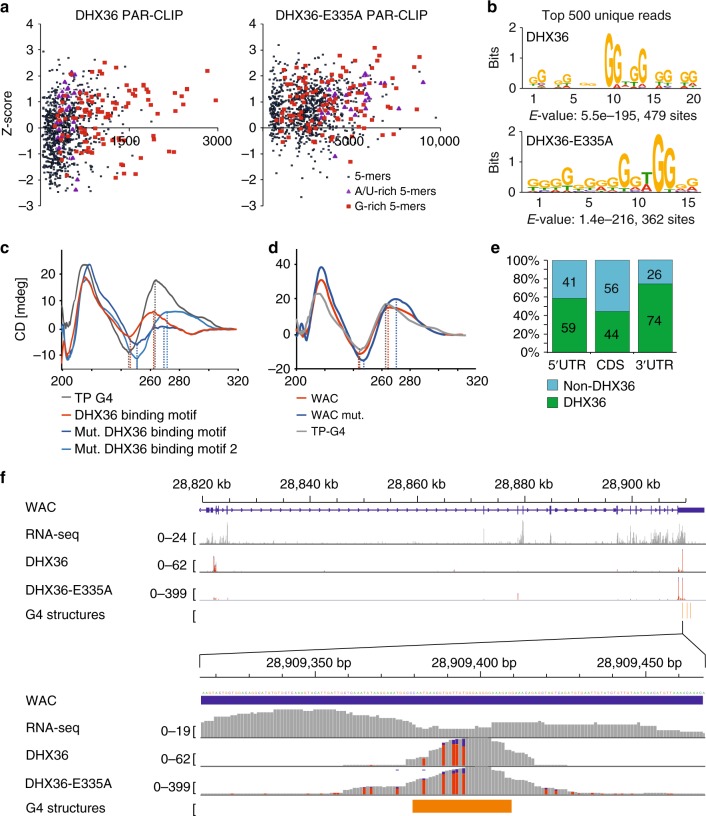
DHX36 recognizes quadruplex-forming G-rich sequence stretches on mRNA. a A comparison of Z-scores and occurrence of all possible 5mers shows an enrichment of G-rich sequences in FH-DHX36 PAR-CLIP binding sites (left panel). Same analysis for FH-DHX36-E335A PAR-CLIP (right panel) shows additional enrichment for A/U-rich 5mers. 5mers containing at least three Gs (red squares) or being A/U-rich (purple triangles) are highlighted. b Weblogo of the RNA recognition element of FH-DHX36 (top) and FH-DHX36-E335A (bottom) PAR-CLIP binding sites generated by MEME (P-value <0.0001) using the top 500 unique reads. c Circular dichroism spectra of oligonucleotides (Supplementary Table 1) after performing a G4-folding protocol. The FH-DHX36 PAR-CLIP-derived RRE (red) shifts towards peaks of positive control TP-G4 (gray), whereas two mutated binding motifs (light and dark blue) do not shift. Lines represent mean of ten subsequent measurements. d Same as c but with a native DHX36 RRE of the WAC mRNA (red) and a mutated version (blue) (Supplementary Table 1). Source data are provided as a Source Data file. e Percent of sites in the human transcriptome forming rG4s in vitro10 categorized by 5′ UTR, CDS, and 3′ UTR found in DHX36 PAR-CLIP binding sites (green). f Top panel: screenshot of the FH-DHX36 and FH-DHX36-E335A PAR-CLIP binding sites for the representative target mRNA WAC. The gene structure is shown, as well as coverage from a HEK293 RNA-seq experiment. The bottom two tracks show the alignment of sequence reads with characteristic T-to-C mutations from a FH-DHX36 and FH-DHX36-E335A PAR-CLIP experiment. Bottom panel: close-up of the indicated region in the 3′ UTR of WAC. rG4s formed in vitro10 are indicated in orange