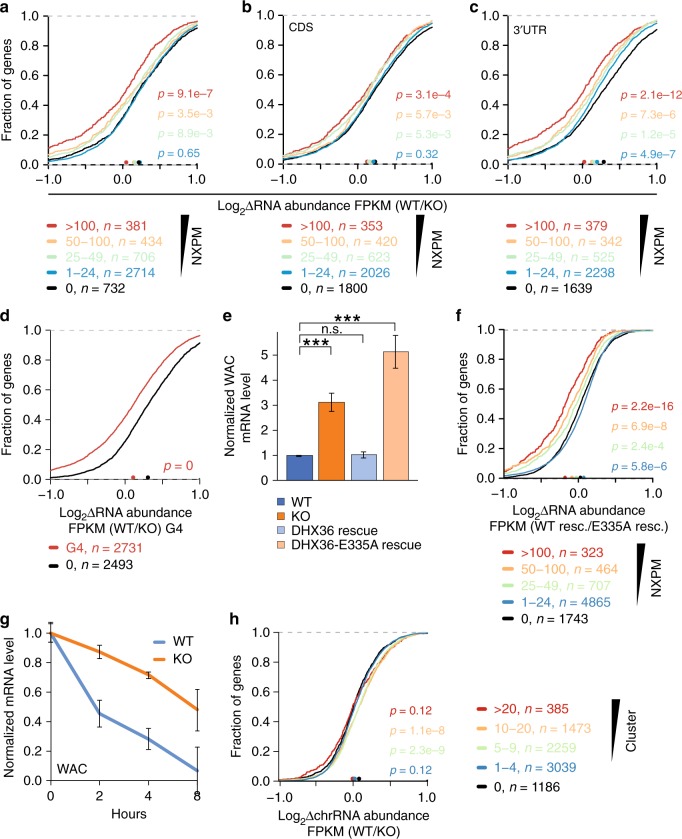
Fig. 4.
DHX36-KO results in increased target RNA abundance. a DHX36-KO results in an increased target mRNA abundance shown by CDF comparing changes in target mRNA abundance of DHX36-KO (n = 3) and parental HEK293 cells (n = 3). Target mRNAs were binned in accordance to the number of NXPM obtained by DHX36-E335A PAR-CLIP. Significance was determined using a two-sided Kolmogorov–Smirnov (KS) test. b Same as in a, except mRNAs were binned based on the number of NXPM in the CDS. c Same as in a, except mRNAs were binned based on the number of NXPM in the 3′ UTR. d Same as in a, except mRNAs were binned based on whether they harbor a G4-site identified previously10 overlapping with PAR-CLIP binding sites. e Quantification of WAC mRNA levels in HEK293 wild-type cells (WT), DHX36-KO cells (KO), DHX36-KO cells with FH-DHX36 overexpression (DHX36 rescue), and DHX36-KO cells with FH-DHX36-E335A overexpression (DHX36-E335A rescue) by RT-qPCR. WAC mRNA levels were normalized to the level of U6 snRNA. Median WAC WT levels were scaled to 1. Significance was calculated using a Student’s t-test (n = 3). Significance levels: *P < 0.05, **P < 0.01, and ***P < 0.001 compared to normalized WT WAC level. Error bars represent standard deviations of three experiments. f Same as in a, except target mRNA abundance in HEK293 DHX36-KO cells with FH-DHX36 overexpression were plotted over target mRNA abundance in HEK293 DHX36-KO cells with FH-DHX36-E335A overexpression. g DHX36 target mRNA WAC show increased half-life upon DHX36-KO shown by qPCR after transcriptional block with actinomycin D and isolation of RNA at the indicated timepoints. Error bars represent standard deviations of three experiments. h DHX36 regulates target mRNA abundance at a posttranscriptional rather than transcriptional level shown by CDFs comparing changes in nascent target mRNA abundance purified from chromatin of DHX36 knockout cells (n = 3) and parental HEK293 cells (n = 3). Target mRNAs were binned in accordance to the number of binding sites obtained by DHX36-E335A PAR-CLIP. Significance was determined using a two-sided KS test. Source data are provided as a Source Data file