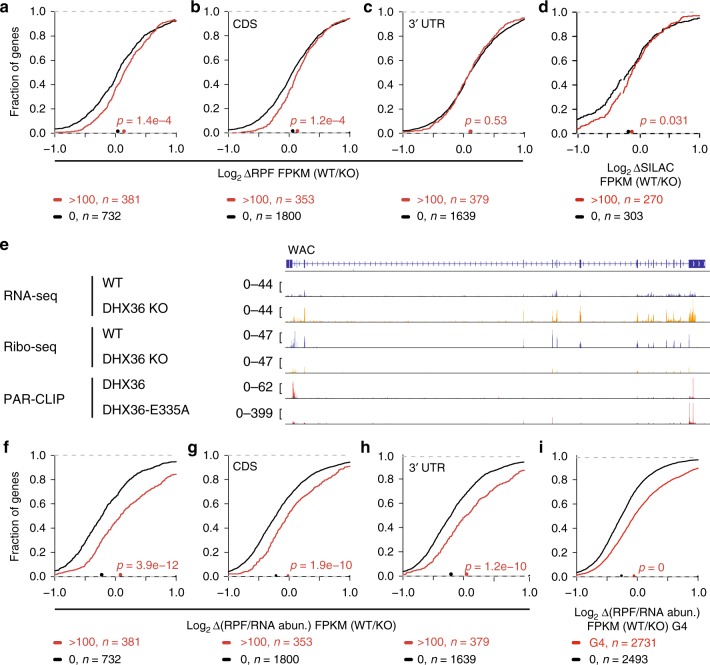
Fig. 5.
DHX36-KO results in reduced target mRNA translation efficiency. a Cumulative distribution function comparing changes in ribosome-protected fragments (RPFs) of DHX36-KO (n = 3) and parental HEK293 cells (n = 3). Target mRNAs are binned in accordance to the number of NXPM obtained by DHX36-E335A PAR-CLIP. Significance was determined using a two-sided KS test. b Same as in a, except target mRNAs were binned in accordance to NXPM in the CDS. c Same as in a, except mRNAs were binned based on the number of NXPM in the 3′ UTR. d Same as in a, except protein abundance changes as determined by SILAC were plotted. e Screenshot of RNA-seq and Ribo-seq coverage in wild-type and DHX36-KO HEK293 cells on the representative DHX36 target WAC. Bottom two tracks show the coverage for DHX36 and DHX36-E335A PAR-CLIP. f Cumulative distribution function comparing changes in translation efficiency (TE, RPF/RNA abundance) of DHX36-KO (n = 3) and parental HEK293 cells (n = 3). Target mRNAs are binned in accordance to the number of NXPM obtained by DHX36-E335A PAR-CLIP. Significance was determined using a two-sided KS test. g Same as in f, except mRNAs were binned based on the number of NXPM in the CDS. h Same as in f, except mRNAs were binned based on the number of NXPM in the 3′ UTR. i Same as in f, except mRNAs were binned based on whether they harbor a G4-site identified previously10 overlapping with PAR-CLIP binding sites or not