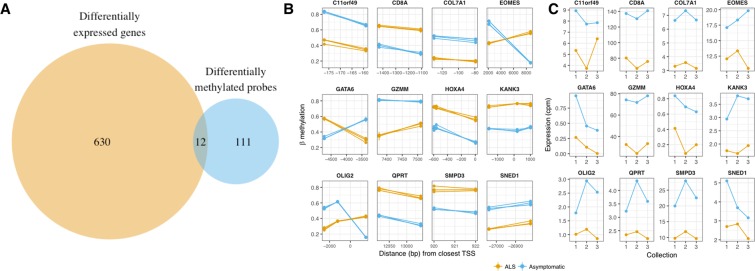
Figure 9.
Twelve overlapping genes were identified in the male SALS twins DMPs and DEGs. (A) While 506 genes were identified as having multiple probes with a difference in β-methylation (≥0.25) between the ALS-discordant male SALS twins, only 123 of these genes were present in the matching RNA-Seq data. 642 of 750 genes identified as differentially expressed were present in the matching DNA methylation data. Twelve of these genes were both differentially expressed and differentially methylated (C11orf49, CD8A, COL7A1, EOMES, GATA6, GZMM, HOXA4, KANK3, OLIG2, QPRT, SMPD3, SNED1). (B) On the HumanMethylation 450K beadchip, multiple CpG probes are annotated to each gene. Methylation of all probes annotated to each of the twelve differentially expressed and differentially methylated genes shows strong consistency within each co-twin. Distance from the transcription start site in base pairs is shown on the x axis. Multiple data points per person at each probe indicate longitudinal sampling. Duplicate collections within a time point are shown as the mean with the standard deviation indicated by a line. (C) Longitudinal expression of the 12 shared genes, (C11orf49, CD8A, COL7A1, EOMES, GATA6, GZMM, HOXA4, KANK3, OLIG2, QPRT, SMPD3, SNED1), is consistently different between ALS discordant male SALS co-twins over time.