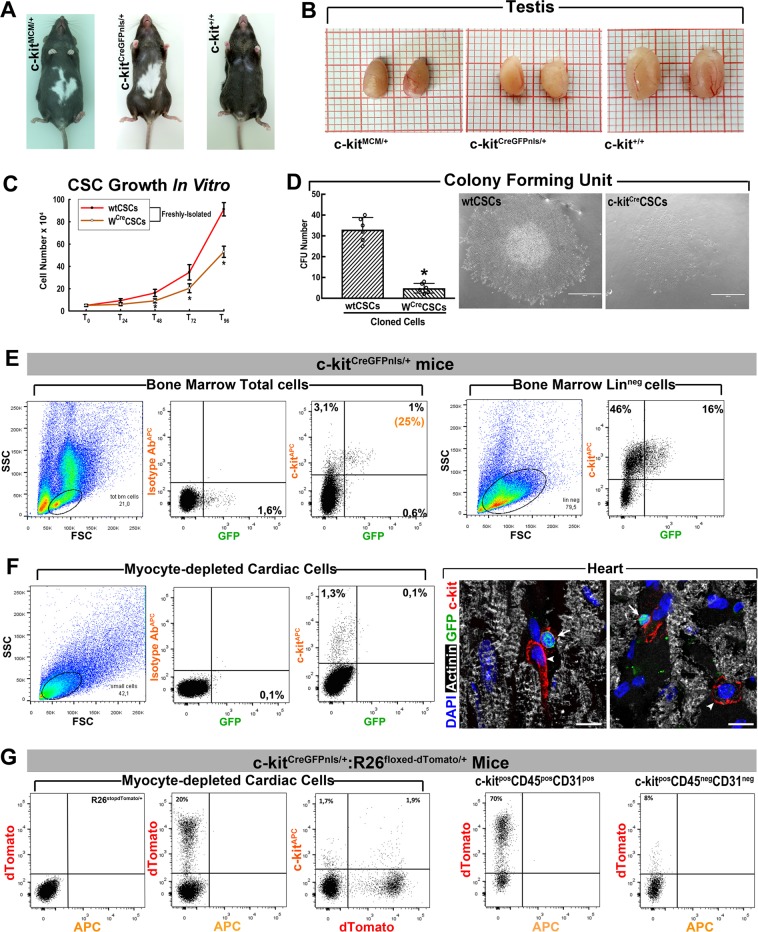
Fig. 4. c-kitCre mice fail to label resident CSCs.
a Photographs of representative adult c-kitMCM and c-kitCreGFPnls/+ mice showing the typical W-mutation-determined piebald and white belly spot as compared with c-kit+/+ C57BL/6J mouse. b The testis from 2-month-old kitMCM and c-kitCreGFPnls/+ show a significant size reduction compared to c-kit+/+ mice. c Cell growth curve of freshly isolated (P1) WCreCSCs vs. wtCSCs over 96 h. n = 7 biological replicates, *p < 0.05 vs. wtCSCs (Kruskal–Wallis test). d Colony number of 100 cloned wtCSCs vs. WCreCSCs in Methocult. n = 7 biological replicates, *p < 0.05 vs. wtCSCs (Student’s t-test). On the right, representative light microscopy images of a colony from wtCSCs and WCreCSCs, respectively. n = 6 plots biological replicates. Scale bars = 1000 µm. e GFP expression (from the c-kitCreGFPnls allele) in the monocyte–lymphocyte gate of total bone marrow (BM) cells population and in total c-kitpos BM cells from 2-month-old c-kitCreGFPnls/+ mice. Less than 25% of the total c-kitpos BM cells express GFP in c-kitCreGFPnls/+ mice. Concurrently, <35% of the lineage-negative (Linneg) c-kitpos BM cells (including HSCs) express GFP in c-kitCreGFPnls/+ mice. f GFP expression (from the c-kitCreGFPnls allele) in cardiomyocyte-depleted cardiac cells (left) and in cardiac c-kitpos cardiac cells (right) from 2-month-old c-kitCreGFPnls/+ mice. Less than 10% of these cells express GFP. Right, confocal microscopy representative images of cardiac cross-sections showing that GFP nuclear expression is expressed only in some (20 ± 3%) of the c-kit-expressing cardiac cells. Scale bar = 20 µm. (e, f representative of n = 5 BM/Hearts). g Mice KI-ed within the first exon of the c-kit locus to express Cre recombinase and GFP with a nuclear localization sequence (eGFPnls) behind an internal ribosome entry site (IRES) (c-kitCreGFPnls or c-kitCre) were crossed with B6.129S6-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J (abbreviated as R26floxed-dTomato) Cre-reporter mice, which harbour a targeted mutation of the Gt(ROSA)26Sor locus with a loxP-flanked STOP cassette preventing transcription of a CAG promoter-driven red fluorescent protein variant (tdTomato), which is expressed following Cre-mediated recombination. Approximately 20% of total cardiac cells and ~60% of c-kitpos cardiac cells from these mice are dTomatopos. Approximately 70% of lineage-committed endothelial/mast cell (CD45pos/CD31posc-kitpos) cardiac cells are recombined to express dTomato, whereas <10% of the CSC-enriched CD45negCD31neg/c-kitpos were recombined to become dTomatopos (g representative of n = 5 hearts). All data are mean ± SD