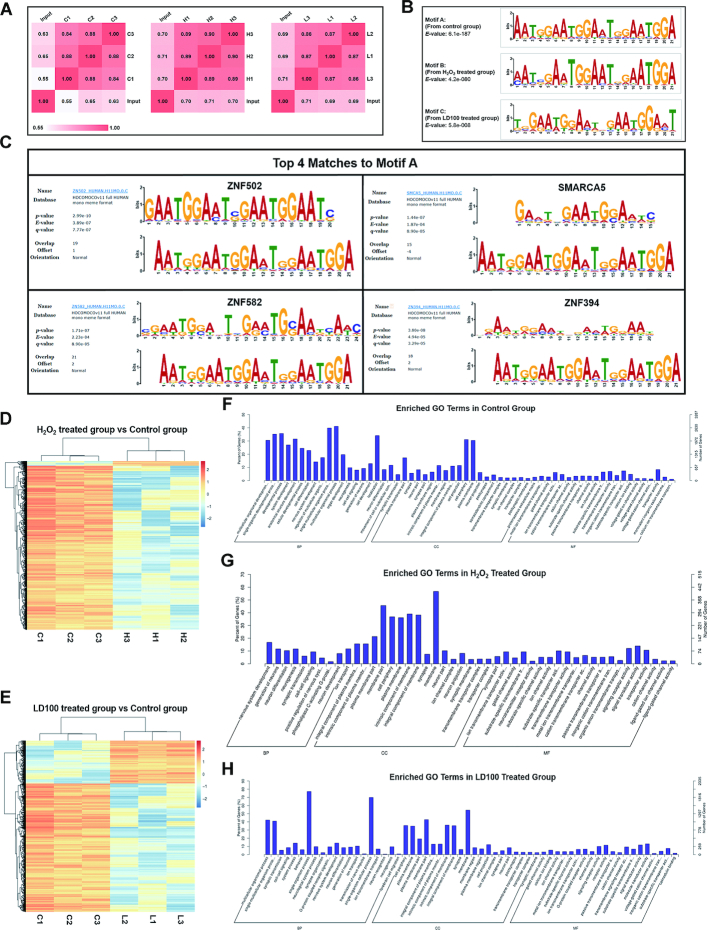
Figure 2.
SOD1 binds to DNA with different sequence preferences under varied redox conditions. (A) Person correlations between parallel ChIP-Seq samples in control (designed as C1, C2 and C3), H2O2 (H1, H2 and H3), and LD100 (L1, L2 and L3) treated groups. Unless otherwise specified, the labels for all ChIP-Seq samples were the same as here. All the ChIP-Seq data were representative of three independent experiments. (B) Typical SOD1 binding motifs under varied redox conditions. (C) Motif logo matching between motif A (bottom) and four consensus motifs (top), determined by TOMTOM (MEME). (D and E) Hierarchical clustering analyses of fold enrichment values in control, H2O2 and LD100 treated groups. (F–H) Enriched GO terms of all ChIP-Seq samples in control (F), H2O2 (G), and LD100 treated groups (H).