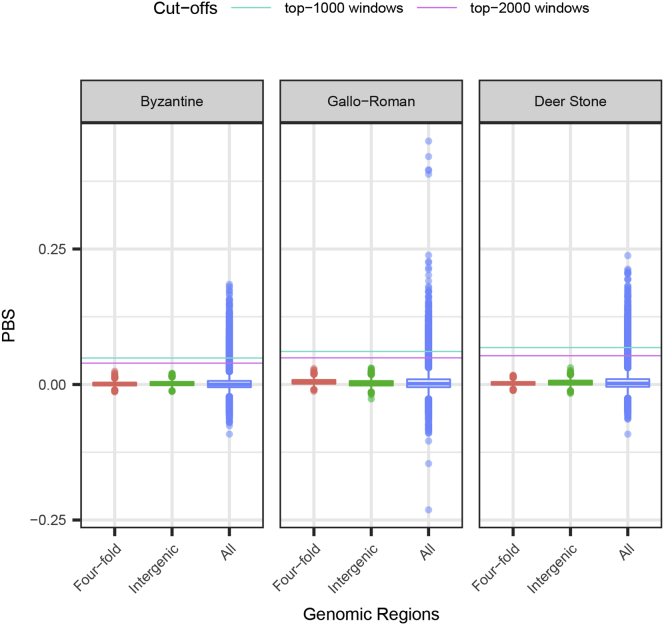
Figure S4.
Distributions of PBS Scores, Related to Figure 4
We considered a 3-population tree consisting of 11 Bronze Age Deer Stone horses (representing the pre-C7th–C9th Asian group), 11 Gallo-Roman horses (pre-C7th–C9th European horses) and 17 Byzantine horses (post-C7th–C9th). All individual genomes showed above 1-fold depth-of-coverage (Tables S6 and S7; STAR Methods). The genome-wide distributions of PBS scores in all three branches within 50kb sliding windows is shown in blue. Neutral distributions are calculated from 50,000 bootstrap pseudo-replicates at four-fold degenerate sites (red) and intergenic positions defined to be located at least five kilobases away from gene bodies (green). Conservative cut-offs retained to identify selection candidates are shown as horizontal lines.